2. 南京师范大学环境学院,江苏 南京 210023
2. School of the Environment, Nanjing Normal University, Nanjing, Jiangsu 210023, China
环境内分泌干扰物(EDCs)又名环境激素,具有类似激素的结构和效应,能够通过模拟内源性激素干扰鱼类、野生动物和人类的内分泌系统[1-2],导致体内稳态失衡、生殖和发育失调、免疫功能改变等不良反应[3-5]。EDCs包含的物质多种多样,大致可分为多氯联苯类(PCBs)、酞酸酯类(PAEs)、酚类和雌激素类,其中酚类EDCs因在工业生产中大量使用,且拟雌激素活性较强,受到广泛关注。酚类EDCs主要包括烷基酚(APs)、双酚类(BPs)和氯酚(CPs)。
APs包括从甲酚到十二烷基酚的许多化合物,其由一个苯酚环组成,苯酚环由可变长度的烷基链单取代或多取代。APs及其聚氧基衍生物作为中间体或添加剂被广泛应用于乳化剂、洗涤剂、浮选和分散剂等工业产品生产中。APs中最具代表性的是壬基酚(NP)和辛基酚(OP),它们不仅具有内分泌干扰性,还具有难降解、环境持久性、远距离运输和生物积累的特性,在低浓度情况下可通过生物积累对内分泌系统产生不良影响,如影响鱼的雌性化作用,与人血清蛋白结合,以及与不同癌症之间存在正相关[6-7],已被联合国环境规划署(UNEP)纳入了27种优先控制持久性毒性污染物(PTS)名单。APs及其前体物的广泛使用导致其在各种水体中普遍存在,其质量浓度从地表水中的644 μg/L到未经处理废水中的1 350 μg/L不等[7],并且已在世界各地许多河流中检测到APs的积累,如瑞士的格拉特河[8],西班牙的埃布罗河[9]和勒布雷加特河[10]等。
BPs含有2个酚基,由碳原子或硫原子相连接,常用于聚碳酸脂、环氧树脂等高分子材料的加工生产中。双酚A(BPA)是典型的BPs,广泛应用于生产阻燃剂、聚碳酸脂和环氧树脂塑料,经常出现在日常生活用品中,包括玩具、食品包装、医疗设备以及电子产品等[11]。BPA能够干扰人和动物的内分泌系统,并干扰人体糖代谢途径和基因表达,对健康造成不良影响[12]。由于BPA在环境中无处不在并威胁人类和动物健康,欧盟、美国和加拿大已禁止BPA用于生产包括婴儿奶瓶在内的儿童护理产品[13]。为了应对这些限制,行业不断开发和使用BPA化学类似物来替代BPA,如双酚S(BPS)、双酚F(BPF)和双酚AF(BPAF)等。然而,这些BPA类似物也存在一定毒性和内分泌干扰效应。由于BPs的广泛使用和与人类的密切接触,近年来,已经在地表水、沉积物、工业废水[14]等环境样本以及人类尿液中检测到BPs的存在。在对亚洲地表水中BPs的调查中发现,BPF是日本、韩国和中国河流中的主要污染物,并且在东京田川河中发现BPF的质量浓度高达2 850 ng/L[15]。对美国、日本和韩国工业化地区沉积物中BPs的调查发现,沉积物中BPs总质量分数为低于定量限值~25 300 ng/g(干重),平均值为201 ng/g(干重)[16]。此外,在人类尿液中BPA的检出率高达92.6%[17]。
CPs是一种氯化芳香族化合物,包括一氯酚(MCPs)、二氯酚(DCPs)、三氯酚(TCPs)、四氯酚(TeCP)和五氯酚(PCP),化学性质稳定,难以生物降解,具有环境持久性,易于通过食物链在生物体内富集,同时具有致畸、致癌、致突变的“三致”作用和内分泌干扰性,是公认的有毒有害污染物。美国国家环境保护局(US EPA)和中国环境监测总站都将多种CPs列为优先控制污染物[18]。CPs常被用做杀菌剂、杀虫剂、除草剂、除锈剂、木材防腐剂,以及作为染料和药品生产的中间体。此外,在饮用水氯化消毒、造纸生产、焦化工艺、木材蒸馏消毒、杀虫剂和除草剂生物降解和有机物燃烧过程中也会产生CPs[19]。CPs广泛存在于各类废水、污泥、地表水和地下水中[20]。Gao等[21]研究发现,2,4-二氯酚(2,4-DCP)和2,4,6-三氯酚(2,4,6-TCP)在黄河、淮河和海河流域的污染尤为严重,而PCP污染主要发生在长江流域,且相较于2,4-DCP和2,4,6-TCP,PCP检出浓度和检出率更高,污染最为严重。韩国某核电站附近的海洋沉积物中CPs的质量分数达到了0.145~16.1 ng/g(干重) [22]。希腊Thermaikos海湾和Loudia河沉积物中均检出了2,4-DCP[23]。一些生活在沉积物中的生物如蛤蜊,其体内PCP质量分数高达133 mg/kg[24]。地表水中氯酚的质量浓度为2~2 000 ng/L[25],并且有研究表明在人体血液和尿液中都发现了PCP[26]。
微生物降解是降低环境中酚类EDCs毒理学风险的有效途径,具有成本低廉、环境友好、去除效率高等特点。近年来,陆续有研究者从受污染的土壤、沉积物和污水处理厂污泥中分离出能够以酚类EDCs为单一碳源和能源进行代谢降解的降解菌。当酚类EDCs存在时,这些降解菌会表达特定的功能基因,产生特定的降解酶来进行酚类EDCs代谢。本文系统综述了好氧和厌氧条件下降解酚类EDCs的功能菌及其降解途径,总结了微生物降解酚类EDCs的功能酶和功能基因,以期为酚类EDCs降解的微生物资源挖掘及微生物控制方法的优化和创新提供支撑。
1 微生物好氧降解酚类EDCs好氧微生物降解是酚类EDCs去除的主要途径,包括APs、BPs、CPs在内的酚类EDCs均已被报道可被多种好氧真菌和细菌降解。这些降解菌可作为酚类EDCs环境控制和修复的潜在可选择性资源,其主要来源于受污染的土壤、河流或海洋沉积物以及活性污泥,其特定降解过程通过相应功能基因表达的酶介导控制。现对其降解菌、功能基因、菌株来源和降解效率进行了总结,见表 1。
| 表 1 酚类EDCs好氧降解菌相关信息① |
APs的微生物好氧降解主要聚焦在NP和OP 2种代表性物质的研究上,在土壤、沉积物、污水和地表水环境中均存在能够对NP和OP进行高效好氧降解的功能菌,包括假单胞菌(Pseudomonas)、寡养单胞菌(Stenotrophomonas)、鞘氨醇菌(Sphingobium)、根瘤菌(Rhizobium)和鞘氨醇单胞菌(Sphingomonas)等细菌菌属,以及简青霉菌(Penicillium simplicissimum)、扩展青霉(Penicillium expansum)、塔宾曲霉(Aspergillus tubingensis)、深黄伞形霉(Umbelopsis isabellina)和镰状镰孢菌(Fusarium falciforme)等真菌(表 1)。不同功能菌的降解效率不尽相同,其中1×108 CFU/g(每克样本中的微生物数量)的鞘氨醇菌Y2(Sphingobium strain Y2)可以在10 d左右的较短时间内将沉积物中质量分数为150 mg/kg的NP和OP几乎完全降解,而1×109 CFU/g的根瘤菌WZ1(Rhizobium strain WZ1)在17 d左右可以将沉积物中质量分数为200 mg/kg的NP降解约80%。
细菌好氧降解APs主要通过羟化酶或单加氧酶使APs羟基化产生醇或苯二酚再进行下一步环的裂解,相关功能基因主要有烷烃羟化酶基因(alkB)、单组分单加氧酶基因(sMO)、多组分苯酚羟化酶基因(mPH)、细胞色素氧化酶基因(Cox1、Cox2、Cox3)、糖转化酶基因(Cel7A、Cel7B)、儿茶酚1,2-双加氧酶基因(C12O)和儿茶酚2,3-双加氧酶基因(C23O)。APs好氧降解途径见图 1。由图 1可见,alkB负责氧化烷基链;sMO可以通过Ⅱ型芳香取代反应(ipso)生成苯二酚和支化醇;mPH可以将已存在的羟基附近的C原子单羟基化[60],在短链羟基酚或中链烷基酚中产生醇和邻苯二酚;Cox1、Cox2、Cox3属于细胞色素P450系统,可以催化芳香族化合物的羟基化、氧化和脱羧;纤维素酶7A(Cel7A)和纤维素酶7B(Cel7B)是糖苷水解酶家族的成员,可以产生将多糖转化为糖的酶[30],而C12O(邻位裂解途径)和C23O(间位裂解途径)负责进一步将芳烃裂解开环[35, 37, 43]。Bai等[61]分离了主要由鞘氨醇单孢菌(Sphingomonas)、假单胞菌(Pseudomonas)、嗜脂环物菌属(Alicycliphilus)和食酸菌属(Acidovorax)组成的能够降解NP的兼性微生物菌团(NP-M2),该菌团具有较高的降解效率,能够在48 h和8 d内将1 000 mg/L的NP分别降解75.61%和89.75%;且中间体的检测及聚合酶链式反应(PCR)分析表明,NP的完全去除是由芳香环的氧化开始的,同时功能基因sMO、mPH和C12O在菌团中检出,参与了NP代谢中苯环的氧化和进一步降解。关于OP降解的功能基因,在菌株Sphingomonas sp.strain PWE1中发现了一种负责编码黄素单加氧酶的基因opdA具有OP降解活性,通过大肠杆菌亚克隆表达opdA能够导致OP消失,并且生成符合Ⅱ型ipso取代反应的代谢物,与该菌属降解NP的机制相同,辛基酚降解酶A(opdA)基因参与了观察到的Ⅱ型ipso取代反应,负责OP生物降解的第一步[37]。
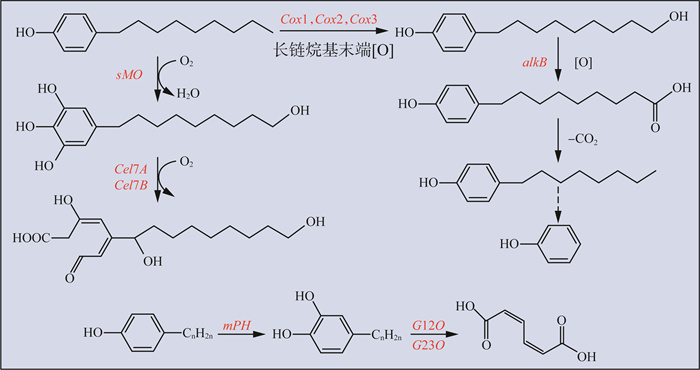
|
图 1 APs好氧降解途径 |
目前,已有研究报道的真菌降解APs的功能酶为漆酶,由WANG等[62]从真菌Ganoderma lucidum纯化得到。漆酶Gl-LAC-4对高浓度的4-正辛基酚(4-n-OP)和APs混合物具有较强的降解能力,并具有对多种金属离子和有机溶剂耐受性强、解毒效果好等特点,对于复杂污染环境中APs的有效处理具有实际应用前景。
1.2 BPs的好氧微生物降解BPs的好氧微生物降解研究主要集中在BPA上,而对BPA的类似物如BPS、BPF、BPB和BPAF的研究相对较少,涉及的功能细菌主要有镰状镰孢菌(Fusarium falciforme RRK20)、恶臭假单胞菌(Pse udomonas putida)、双鞘鞘氨醇菌单胞菌(Sphingomonas bisphenolicum)、贪铜菌(Cupriavidus basilensis)和帕氏假单胞菌(Pseudomonas palleroniana)等(表 1),且这些功能菌均有较好的降解效果,降解效率均>90%(表 1)。
微生物通过单加氧酶将氧引入到BPs环体系来好氧降解BPs,主要的代谢途径是BPs的羟基化[40]。已知的相关功能基因主要为bisdA和bisdB,其负责编码的细胞色素P450在BPs降解的催化羟基化过程中起关键作用[63],参与了降解的第一步[39]。Li等[41]从紫萍根部分离出一株BPA降解菌——新鞘氨醇菌FID3(Novosphingobium sp.FID3),可在12 h可完全降解114 mg/L的BPA,而在细胞色素P450单加氧酶的抑制剂(甲替拉酮)存在时仅能轻微降解BPA(< 10%),证明了细胞色素P450单加氧酶系统对BPA降解的重要性。Badiefar等[40]从石化废水中分离出一株BPA降解菌——格尔格维埃肠杆菌BYK-7(Enterobacter gergoviae strain BYK-7),通过将bisdAB操纵子克隆到E. gergoviae BYK-7中显著提高了E. gergoviae BYK-7的生长和BPA降解活性。
除此之外,应用无标记的定量蛋白质组学研究从活性污泥中分离出的降解菌——鞘氨醇菌BiD32(Sphingobium sp. BiD32)对BPA的代谢,发现负责原儿茶酸降解的基因所编码的蛋白在BPA存在时表达上调,包括与proC、proB、proA和proJ基因同源的蛋白,其中基因proB和proA分别编码原儿茶酸4,5-双加氧酶的小亚基和大亚基,proC编码的蛋白质与其他原儿茶酸降解菌的4-羧基-2-羟基粘康酸-6-半醛脱氢酶(CHMS)基因高度同源,proJ负责编码原儿茶酸降解过程中的4-草酸膦酸水合酶。研究还检测到Sphingobium sp. BiD32中proF可以将BPA转化为简单的含氧官能团,并发现一种新的对羟基苯甲酸酯羟化酶与所检测到的代谢物相关的代谢途径有关[64]。此外,也有报道欧洲亚硝化单胞菌(Nitrosomonas europaea)中的氨单加氧酶[65]和假单胞菌LBC1(Pseudomonas sp. LBC1)中的细胞外漆酶[66]参与BPA的降解,且通过毕赤酵母(Pichia pastoris)过表达来自短芽孢杆菌(Bacillus pumilus)漆酶可用来有效降解BPA[67]。BPA好氧降解途径见图 2。由图 2可见,BisdAB或proF将BPA转化为简单的酚类,再由proABCJ将其羟基化,随后进入三羧酸(TCA)循环。
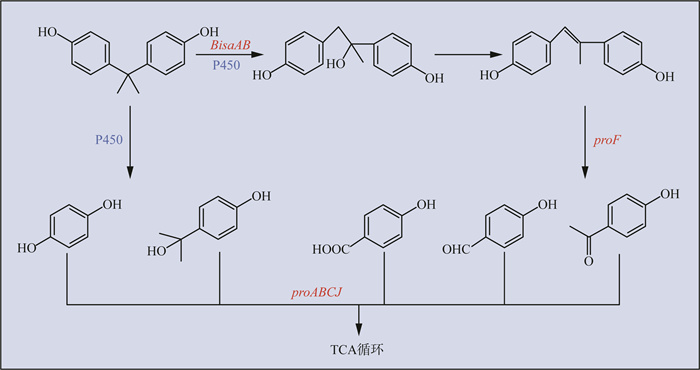
|
图 2 BPA好氧降解途径 注:红色为功能基因,蓝色为功能酶。 |
在BPA类似物的好氧微生物降解研究中,Wang等[68]从河流沉积物中富集得到一个BPS降解菌群,其中生丝微菌属(Hyphomicrobium)、潘多拉菌属(Pandoraea)、红球菌属(Rhodococcus)和贪铜菌属(Cupriavidus)4个菌属为优势菌,在pH值为7和30 ℃条件下,该菌群可在10 d内将初始质量浓度为50 mg/L的BPS降解99%。Yin等[69]采用白腐真菌(Phanerochaete sordida YK-624)在非木质素降解条件下降解BPF,7 d可将0.1 mmol/L BPF完全降解,在此过程中,BPF经细胞色素P450s氧化转化为4,4-二羟基二苯甲酮,并进一步转化为4-羟基苯甲酸。而白腐真菌(Phanerochaete sordida YK-624)在木质素降解条件下,4 d内可将0.1 mmol/L BPF完全降解,降解过程中较高水平的木质素分解酶(木质素过氧化物酶和锰过氧化物酶)发挥了重要作用,而细胞色素P450s作用较小[70]。因此,可降解BPF的木质素分解酶和细胞色素P450s的降解作用因降解条件而异。
1.3 CPs的好氧微生物降解CPs好氧微生物降解的研究对象包括MCPs、DCPs和多氯酚,相关降解菌主要包括氯酚节杆菌A6(Arthrobacter chlorophenolicus A6)、混浊红球菌(Rhodococcus opacus)、富养罗尔斯通氏菌JMP-134(Ralstonia eutropha JMP134)、洋葱伯克霍尔德氏菌AC1100(Burkholderia cepacia AC1100)、假单胞菌属(Pseudomonas strains)、固氮菌GP1(Azotobacter sp. strain)、门多萨假单胞菌(Pseudomonas mendocina)、类诺卡氏菌PD653(Nocardioides sp. strain PD653)、氯酚鞘氨醇菌ATCC 39723(Sphingomo nas chlorophenolica ATCC39723) 等,大部分功能菌降解效率>90%。
微生物降解CPs主要涉及加氧酶和双加氧酶。微生物好氧降解MCPs和DCPs,由于其氯基团数量较少,可以在不事先脱氯的情况下实现环的裂解和降解,能够使邻位羟基化的单加氧酶[如4-CP-2-单加氧酶(tfdB)、2-CP-6-单加氧酶和2,4-DCP-6-单加氧酶(tfdA)]负责将MCPs和DCPs初步转化为氯儿茶酚(CCs)。另外,具有氧化脱氯功能的4-CP-4-单加氧酶也能够参与4-氯酚(4-CP)好氧微生物降解的第一步,产生无氯的对苯二酚和偏苯三酚,氯酚降解基因cphA-Ⅰ和cphA-Ⅱ2个基因是氯酚降解基因簇(cph基因)的一部分,负责编码功能性偏苯三酚1,2-双加氧酶[48-49][图 3(a)]。单加氧酶催化CPs完成第一步降解后,得到双羟基化而失去芳香性的CCs或无氯的对苯二酚和偏苯三酚,具有环裂解功能的双加氧酶开始发挥作用,进行环裂解途径。需氧环裂解可以通过儿茶酚2,3-双加氧酶(C23O)参与的间位裂解途径[71]或儿茶酚1,2-双加氧酶(C12O)参与的邻位裂解途径实现;邻位裂解途径存在3种类型,即通过邻苯二酚降解非氯化芳烃的邻位Ⅱ途径,通过氯邻苯二酚降解氯化芳烃的一种修饰的邻位型Ⅱ途径[72],以及新修饰的修饰邻位裂解途径,该途径中氯邻苯二酚1,2-双加氧酶(tfdC)具有高度活性。
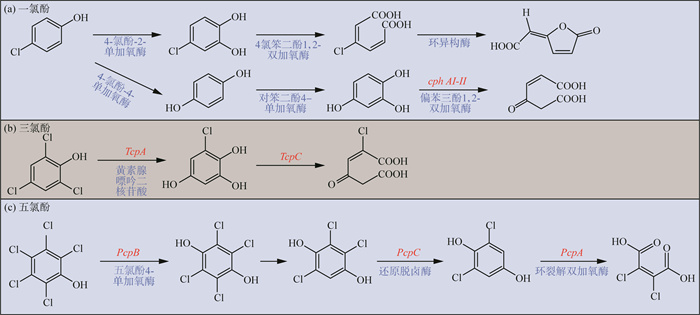
|
图 3 CPs好氧降解途径 注:红色为功能基因,蓝色为功能酶。 |
多氯酚由于有更多的氯基团,比MCPs和DCPs更具毒性和难降解性,不能在不事先脱氯的情况下实现环的裂解和降解,因此,脱氯是多氯酚矿化的第一步。富养罗尔斯通氏菌(Ralstonia eutropha JMP134)降解2,4,6-TCP通过初步产生氯对苯二酚再进一步进行好氧生物降解,功能基因tcp编码功能酶,包括参与TCP初步脱氯的2,4,6-TCP单加氧酶(tcpA)、负责环裂解的6-氯羟基醌1,2-双加氧酶(tcpC)以及马来乳酸还原酶(tcpD)[52-53, 73][图 3(b)]。洋葱伯克霍尔德氏菌(Burkholderia cepacia AC1100)降解2,4,5-TCP通过一种依赖于黄素腺嘌呤二核苷酸(FADH2)的2,4,5-TCP 4单加氧酶(TftD)氧化还原将其转化为5-氯-2-羟基对苯二酚,反应过程中的FADH2由黄素还原酶(TftC)提供[54]。类诺卡氏菌(Nocardioides sp. strain PD653)降解PCP过程中发现了新的功能基因hcbB1-3,与TftD功能相似[58]。氯酚鞘氨醇菌(Sphingomonas chlorophenolica ATCC39723)降解PCP由pcpB基因编码的PCP 4-单加氧酶(pcpB)转化为四氯对苯二酚(TeCHQ),随后TeCHQ还原脱氯为三氯对苯二酚(TrCHQ),然后通过由pcpC基因编码的还原脱卤酶转化为2,6-二氯对苯二酚(2,6-DCHQ),新型环裂解双加氧酶PcpA将2,6-DCHQ转化为2-氯马来酸[59][图 3(c)]。除此之外,细胞色素P450型加氧酶也参与了多氯酚羟基化过程[74-75]。
2 微生物厌氧降解酚类EDCs厌氧微生物降解APs和BPs的相关研究相较于好氧较少,且降解效率较低。而缺氧或厌氧条件下进行微生物还原脱氯是CPs一个重要的去除途径。现对其降解菌、功能基因、菌株来源和降解效率进行了总结,见表 2。
| 表 2 酚类EDCs厌氧降解菌相关信息 |
厌氧条件下微生物单菌或菌群降解APs的研究相对于好氧条件下较少,降解效率较低,降解时间较长。APs厌氧降解菌主要包括丙酸杆菌属(Propionibacterium)、嗜蛋白质菌属(Proteiniphilum)、伽玛变形菌纲(Gammaproteobacteria)、德尔塔变形菌纲(Deltaproteobacteria)、绿弯菌纲(Choloroflexi)和厚壁菌门(Firmicutes)等,降解效率普遍低于好氧条件下的功能菌降解(表 2)。在硝酸盐或硫酸盐还原条件下,变形菌门(Proteobacteria)、厚壁菌门(Firmicutes)、拟杆菌门(Bacteroidetes)和绿屈挠菌门(Chloroflexi)菌属在NP生物降解过程中占优势[77, 82]。
有研究发现厌氧发酵生产过程中,添加表面活性剂和碱性pH条件可以提高NP的生物降解,pH值为10.0和表面活性剂聚环氧乙烯月桂酰醚(Brij 35)存在时,特异性NP降解菌的丰度以及功能基因烷烃羟化酶基因(alkB)和单组分单加氧酶基因(sMO)的数量增多,有利于NP的生物降解,且发酵系统中的产酸细菌可以通过提供底物和中间体共代谢NP增强NP的生物降解[76]。河流沉积物和污泥中厌氧微生物对NP的降解速率从高到低的顺序为硫酸盐还原条件>产甲烷条件>硝酸盐还原条件[83-84]。产甲烷条件下乙酸激酶(ackA)参与了NP和OP的转化,只有含有羧基或羟基并具有适度空间位阻的化合物才会被该酶转化,且较大可能是通过磷酸化进行转化。此外,一些候选酶,如丙酸激酶(ppk)和丁酸激酶(buk),也可以进行磷酸化,可能参与NP和OP的进一步转化[85]。
2.2 BPs的厌氧微生物降解关于BPs微生物厌氧降解的研究较少,且降解效果不显著。例如,从河水中分离细菌降解BPA,在好氧条件下,BPA降解半衰期为2~3 d,第10 d BPA质量浓度降到检出限(< 0.005 mg/L),但在厌氧条件下,30 ℃持续10 d几乎没有发现BPA质量浓度的下降[86]。在缺氧河口沉积物降解四氯双酚A、四溴双酚A和BPA的研究中,发现四溴双酚A和四氯双酚A在产甲烷或硫酸盐还原的条件下作为主要终端电子受体发生了脱卤反应,而BPA和脱卤反应产生的BPA没有降解[87]。然而,Yang等[78]研究发现在硝酸盐或硫酸盐还原条件下,河流沉积物中BPA经过120 d的降解率分别达到93%和89%,分析得到变形菌门(Proteobacteria)、拟杆菌门(Bac-teroidetes)、绿屈挠菌门(Chloroflexi)、厚壁菌门(Firmicutes)、出芽单胞菌门(Gemmatimonadetes)和放线菌门(Actinobacteria)是沉积物中主要的菌群(表 2)。
2.3 CPs的厌氧微生物降解CPs作为卤代芳香化合物,在缺氧或厌氧条件下进行微生物还原脱氯是一个重要的生物去除途径。厌氧降解CPs的功能菌主要有Desulfomonile tiedjei、脱亚硫酸杆菌属(Desulfitobacterium)、假单胞菌(Pseudomonas)、脱卤素杆菌属(Dehalobacter strains)和梭菌属(Clostridium)等,主要来源于河流沉积物或污水污泥(表 2),其中Desulfomonile tiedjei是一种严格厌氧的革兰氏阴性硫酸盐还原菌,是厌氧条件下降解CPs的最佳细菌[88-89]。CPs在厌氧降解过程中需要引入氢或辅助底物作有效电子供体,提高其降解速率。氢被广泛用作还原脱氯过程中的电子供体。Li等[79]从被氯化脂肪烃污染的沉积物中富集得到可以邻位还原2,4,6-TCP脱氯的培养物,在15 d内,利用H2和酵母提取物分别作为电子供体和碳源,可将100 mmol/L的2,4,6-三氯酚(2,4,6-TCP)脱氯为4-CP,并证实了培养物中2种新的脱盐杆菌菌株TCP-5和TCP-6参与还原性脱卤过程,并对卤代芳香化合物和脂肪族化合物均具有广泛的有机卤化物利用活性。El-Sayed[90]从尼罗河沉积物得到了一个稳定的还原脱氯2,3-DCP和2,4,6-TCP的无沉淀物厌氧微生物群落。当氢作为电子供体,乙酸盐作为碳源,2,3-DCP和2,4,6-TCP作为电子受体时,具有稳定脱氯作用,并发现脱硫菌是脱氯培养物的主要部分。Song等[91]从活性污泥中驯化出可将2,4,6-TCP邻位脱氯生成4-CP的菌群,驯化过程中硫酸还原菌[脱硫葱头状菌属(Desulfobulbus)、脱硫弧菌属(Desulfovibrio)、脱硫微菌属(Desulfoicrobium)和共养菌属(Syntrophus)]逐渐取代δ-变形菌,硫酸还原菌的出现为脱氯菌群的硫酸盐还原能力提供了微生物学证据。
还原脱卤酶(Rdase)是脱氯菌脱氯过程中关键酶,不同的脱氯菌可能有不同功能基因(如crdA和cprA1-5)负责编码Rdase,并且通过不同的脱氯途径(间位、邻位或对位)脱氯。哥本哈根脱亚硫酸杆菌(Desulfitoba-cterium hafniense PCP-1)中已被鉴定存在2种Rdase,一种是在邻位脱氯系统中发挥作用由crdA基因编码的2,4,6-TCP Rdase;一种是cprA5基因编码的3,5-DCP Rdase,其能在中间和对端位置脱氯[80]。Villemur[81]从产甲烷菌群中分离到一株哥本哈根脱亚硫酸杆菌(D. hafniense PCP-1),对cprA3和cprA5的Rdase活性进行了表征,cprA3参与高氯酚的脱氯,能够表达邻位脱氯活性,并观察到cprA3对3,5-DCP和2,3,5-TCP的间位脱氯活性最高;对PCP、2,3,4,5-TeCP和3,4,5-TCP对位脱氯活性最高,仅观察到其对2,4,6-TCP,2,4,5-TCP和2,4-DCP在邻位的脱氯作用。
3 结论及展望酚类EDCs具有内分泌干扰性,危害动物和人类健康,主要通过生活污水和工业废水进入水环境中。受污染的土壤、河流或海洋沉积物和污水污泥中可以分离出能够降解酚类EDCs的微生物菌群或菌株,根据微生物的降解特性对酚类EDCs进行好氧或厌氧降解。APs和BPs的好氧降解相比厌氧研究更多且降解效率更高,而CPs的好氧降解和厌氧还原脱氯都是重要的生物去除途径。酚类EDCs的好氧降解主要途径是在单加氧酶或羟化酶的作用下羟基化,再在双加氧酶的作用下裂解开环进一步矿化。CPs的还原脱氯则是在还原脱卤酶的作用下,以H2作为电子供体实现脱氯。
相比于物理和化学技术,生物去除技术具有经济和环境友好的优势。本文涉及的酚类EDCs降解功能菌是进行污染土壤生物原位修复以及废水中酚类EDCs去除的有力工具。此外,总结了目前已知的参与酚类EDCs降解的功能基因和功能酶,可以在未来研究中作为酚类EDCs环境污染的生物标志物,也可利用这些已知降解功能基因对其他菌株进行基因调控,提高菌株的降解活性。一些研究发现,菌群比单独菌株具有更高的污染物降解效率,可能是存在代谢互作机制或是功能基因、功能酶互补。因此,明晰功能菌、功能基因和功能酶在酚类EDCs降解转化过程中的作用机制,有助于指导性酚类EDCs高效降解菌(群)的定向构建,实现酚类EDCs污染的高效控制。
| [1] |
HOTCHKISS A K, RIDER C V, BLYSTONE C R, et al. Fifteen years after "Wingspread"—environmental endocrine disrupters and human and wildlife health: Where we are today and where we need to go[J]. Toxicological Sciences, 2008, 105(2): 235-259. DOI:10.1093/toxsci/kfn030 |
| [2] |
TYLER C, JOBLING S, SUMPTER J. Endocrine disruption in wildlife: A critical review of the evidence[J]. Critical Reviews In Toxicology, 1998, 28(4): 319-361. DOI:10.1080/10408449891344236 |
| [3] |
COLBORN T, VOM SAAL F S, SOTO A M. Developmental effects of endocrine-disrupting chemicals in wildlife and humans[J]. Environmental Health Perspectives, 1993, 101(5): 378-384. DOI:10.1289/ehp.93101378 |
| [4] |
SAFEA S. Endocrine disruptors and human health: Is there a problem[J]. Toxicology, 2004, 205(1-2): 3-10. DOI:10.1016/j.tox.2004.06.032 |
| [5] |
ANNAMALAI J, NAMASIVAYAM V. Endocrine disrupting chemicals in the atmosphere: Their effects on humans and wild-life[J]. Environment International, 2015, 76(Mar.): 78-97. |
| [6] |
BECHAMBI O, NAJJAR W, SAYADI S. The nonylphenol degradation under UV irradiation in the presence of Ag-ZnO nanorods: Effect of parameters and degradation pathway[J]. Journal of the Taiwan Institute of Chemical Engineers, 2016, 60: 496-501. DOI:10.1016/j.jtice.2015.11.017 |
| [7] |
BHANDARI G, BAGHERI A R, BHATT P, et al. Occurrence, potential ecological risks, and degradation of endocrine disrupter, nonylphenol, from the aqueous environment[J]. Chemosphere, 2021, 275(Jul.): 130013.1-16. |
| [8] |
VOUTSA D, HARTMANN P, SCHAFFNER C, et al. Benzotriazoles, alkylphenols and bisphenol A in municipal waste-waters and in the Glatt River, Switzerland[J]. Environmental Science and Pollution Research, 2006, 13(5): 333-341. DOI:10.1065/espr2006.01.295 |
| [9] |
LACORTE S, RALDŲ A D, MARTÍNEZ E, et al. Pilot survey of a broad range of priority pollutants in sediment and fish from the Ebro river basin(NE Spain)[J]. Environmental Pollution, 2006, 140(3): 471-482. DOI:10.1016/j.envpol.2005.08.008 |
| [10] |
PETROVIC M, SOLÉ M, LÓPEZ DE ALDA M J, et al. Endocrine disruptors in sewage treatment plants, receiving river waters, and sediments: Integration of chemical analysis and biological effects on feral carp[J]. Environmental Toxicology and Chemistry: An International Journal, 2002, 21(10): 2146-2156. |
| [11] |
VANDENBERG L N, HAUSER R, MARCUS M, et al. Human exposure to bisphenol A(BPA)[J]. Reproductive Toxicology, 2007, 24(2): 139-177. DOI:10.1016/j.reprotox.2007.07.010 |
| [12] |
YUE S, YU J, KONG Y, et al. Metabolomic modulations of HepG2 cells exposed to bisphenol analogues[J]. Environment International, 2019, 129: 59-67. DOI:10.1016/j.envint.2019.05.008 |
| [13] |
HERCOG K, MAISANABA S, FILIPIC M, et al. Genotoxic activity of bisphenol A and its analogues bisphenol S, bisphenol F and bisphenol AF and their mixtures in human hepatocellular carcinoma(HepG2) cells[J]. Science of The Total Environment, 2019, 687: 267-276. DOI:10.1016/j.scitotenv.2019.05.486 |
| [14] |
LEE S, LIAO C, SONG G J, et al. Emission of bisphenol analo-gues including bisphenol A and bisphenol F from wastewater treatment plants in Korea[J]. Chemosphere, 2015, 119: 1000-1006. DOI:10.1016/j.chemosphere.2014.09.011 |
| [15] |
YAMAZAKI E, YAMASHITA N, TANIYASU S, et al. Bisphenol A and other bisphenol analogues including BPS and BPF in surface water samples from Japan, China, Korea and India[J]. Ecotoxicology and Environmental Safety, 2015, 122(Dec.): 565-572. |
| [16] |
LIAO C Y, LIU F, MOON H B, et al. Bisphenol analogues in sediments from industrialized areas in the United States, Japan and Korea: Spatial and temporal distributions[J]. Environmental Science & Technology, 2012, 46(21): 11558-11565. |
| [17] |
CALAFAT A M, YE X, WONG L Y, et al. Exposure of the US population to bisphenol A and 4-tertiary-octylphenol: 2003—2004[J]. Environmental Health Perspectives, 2008, 116(1): 39-44. DOI:10.1289/ehp.10753 |
| [18] |
LACORTE S, GUIFFARD I, FRAISSE D, et al. Broad spectrum analysis of 109 priority compounds listed in the 76/464/CEE Council Directive using solid-phase extraction and GC/EI/MS[J]. Analytical Chemistry, 2000, 72(7): 1430-1440. DOI:10.1021/ac991080w |
| [19] |
CZAPLICKA M. Sources and transformations of chlorophenols in the natural environment[J]. Science of The Total Environment, 2004, 322(1-3): 21-39. DOI:10.1016/j.scitotenv.2003.09.015 |
| [20] |
OLANIRAN A O, IGBINOSA E O. Chlorophenols and other related derivatives of environmental concern: Properties, distribution and microbial degradation processes[J]. Chemosphere : Environmental Toxicology and Risk Assessment, 2011, 83(10): 1297-1306. |
| [21] |
GAO J, LIU L, LIU X, et al. Levels and spatial distribution of chlorophenols-2, 4-Dichlorophenol, 2, 4, 6-trichlorophenol, and pentachlorophenol in surface water of China[J]. Chemosphere, 2008, 71(6): 1181-1187. DOI:10.1016/j.chemosphere.2007.10.018 |
| [22] |
SIM W J, LEE S H, LEE I S, et al. Distribution and formation of chlorophenols and bromophenols in marine and riverine environments[J]. Chemosphere, 2009, 77(4): 552-558. DOI:10.1016/j.chemosphere.2009.07.006 |
| [23] |
ALBANIS T A, DANIS T G. Analysis of phenolic compounds in soil and sediment by using SPE-disks and gas chromatography techniques[J]. International Journal of Environmental Analytical Chemistry, 1999, 74(1): 55-67. |
| [24] |
GARBA Z N, ZHOU W M, LAWAN I, et al. An overview of chlorophenols as contaminants and their removal from wastewater by adsorption: A review[J]. Journal of Environmental Management, 2019, 241: 59-75. |
| [25] |
CZAPLICKA M. Sources and transformations of chlorophenols in the natural environment[J]. Science of The Total Environment, 2004, 322(1-3): 21-39. DOI:10.1016/j.scitotenv.2003.09.015 |
| [26] |
ATUMA S S, OKOR D I. Gas chromatographic determination of pentachlorophenol in human blood and urine[J]. Bull Environ Contam Toxicol(United States), 1985, 35(3): 406-410. |
| [27] |
ZHAO W, YUYIN Y, WEIMIN S, et al. Variation of nonylphenol-degrading gene abundance and bacterial community structure in bioaugmented sediment microcosm[J]. Environmental Science and Pollution Research International, 2015, 22(3): 2342-2349. DOI:10.1007/s11356-014-3625-x |
| [28] |
WANG Z, YANG Y, SUN W, et al. Biodegradation of nonylphenol by two alphaproteobacterial strains in liquid culture and sediment microcosm[J]. International Biodeterioration & Biodegradation, 2014, 92: 1-5. |
| [29] |
YAN Z, YING L, HAN D, et al. The nonylphenol biodegradation study by estuary sediment-derived fungus Penicillium simplicissimum[J]. Environmental Science and Pollution Research International, 2016, 23(15): 15122-15132. DOI:10.1007/s11356-016-6656-7 |
| [30] |
YANG Z L, SHI Y Q, ZHAN Y, et al. Different pathways for 4-n-nonylphenol biodegradation by two Aspergillus strains derived from estuary sediment: Evidence from metabolites determi-nation and key-gene identification[J]. Journal of Hazardous Materials, 2018, 359(Oct.): 203-212. |
| [31] |
KUZIKOVA I, SAFRONOVA V, ZAYTSEVA T, et al. Fate and effects of nonylphenol in the filamentous fungus Penicillium expansum isolated from the bottom sediments of the Gulf of Finland[J]. Journal of Marine Systems, 2017, 171: 111-119. DOI:10.1016/j.jmarsys.2016.06.003 |
| [32] |
SOARES A, GUIEYSSE B, DELGADO O, et al. Aerobic biodegradation of nonylphenol by cold-adapted bacteria[J]. Biotechnology Letters, 2003, 25(9): 731-738. DOI:10.1023/A:1023466916678 |
| [33] |
TADASHI T, MANABU M, KAZUTAKA K, et al. Acceleration of nonylphenol and 4-tert-octylphenol degradation in sediment by Phragmites australis and associated rhizosphere bacteria[J]. Environmental Science & Technology, 2011, 45(15): 6524-6530. |
| [34] |
KUZIKOVA I, RYBALCHENKO O, KURASHOV E, et al. Defense responses of the marine-derived fungus Aspergillus tubingensis to alkylphenols stress[J]. Water, Air, & Soil Pollution: An International Journal of Environmental Pollution, 2020, 231(1): 271. |
| [35] |
JANICKI T, KRUPINSKI M, DLUGONSKI J. Degradation and toxicity reduction of the endocrine disruptors nonylphenol, 4-tert-octylphenol and 4-cumylphenol by the non-ligninolytic fungus Umbelopsis isabellina[J]. Bioresource Technology, 2016, 200: 223-229. DOI:10.1016/j.biortech.2015.10.034 |
| [36] |
KUZIKOVA I L, MEDVEDEVA N G. Long-chain alkylphenol biodegradation potential of soil ascomycota[J]. Doklady Biological Sciences, 2023, 511(1): 228-234. DOI:10.1134/S0012496623700515 |
| [37] |
PORTER A W, HAY A G. Identification of opdA, a gene involved in biodegradation of the endocrine disrupter octylphenol[J]. Applied and Environmental Microbiology, 2007, 73(22): 7373-7379. DOI:10.1128/AEM.01478-07 |
| [38] |
RAJENDRAN R K, LEE YW, CHOU PH, et al. Biodegradation of the endocrine disrupter 4- t-octylphenol by the non-ligninolytic fungus Fusarium falciforme RRK20: Process optimization, estrogenicity assessment, metabolite identification and proposed pathways[J]. Chemosphere, 2020, 240(C): 124876. |
| [39] |
SASAKI M, TSUCHIDO T, MATSUMURA Y. Molecular cloning and characterization of cytochrome P450 and ferredoxin genes involved in bisphenol A degradation in Sphingomonas bisphenolicum strain AO1[J]. Journal of Applied Microbiology, 2008, 105(4): 1158-1169. DOI:10.1111/j.1365-2672.2008.03843.x |
| [40] |
BADIEFAR L, YAKHCHALI B, RODRIGUEZ-COUTO S, et al. Biodegradation of bisphenol A by the newly-isolated Enterobacter gergoviae strain BYK-7 enhanced using genetic manipula-tion[J]. Rsc Advances, 2015, 5(37): 29563-29572. DOI:10.1039/C5RA01818H |
| [41] |
LI Y, TOYAMA T, FURUYA T, et al. Sustainable biodegradation of bisphenol A by Spirodela polyrhiza in association with Novosphingobium sp. FID3[J]. Journal of Water and Environment Technology, 2014, 12(1): 43-54. DOI:10.2965/jwet.2014.43 |
| [42] |
ADEL E, YANG J, RUTH N, et al. Biodegradation of endocrine disruptor Bisphenol A by Pseudomonas putida strain YC-AE1 isolated from polluted soil, Guangdong, China[J]. BMC Microbiology, 2020, 20(1): 11-25. DOI:10.1186/s12866-020-1699-9 |
| [43] |
KO-ICHI O, YUJI T, TOMOAKI N, et al. Isolation and characterization of a novel bacterium, Sphingomonas bisphenolicum strain AO1, that degrades bisphenol A[J]. Biodegradation, 2007, 18(2): 247-255. DOI:10.1007/s10532-006-9059-5 |
| [44] |
LOBOS J H, TERRY K L, SU T M. Biodegradation of bisphenol A and other bisphenols by a gram-negative aerobic bacterium[J]. Applied and Environmental Microbiology, 1992, 58(6): 1823-1831. DOI:10.1128/aem.58.6.1823-1831.1992 |
| [45] |
FISCHER J, KAPPELMEYER U, KASTNER M, et al. The degradation of bisphenol A by the newly isolated bacterium Cupriavidus basilensis JF1 can be enhanced by biostimulation with ph-enol[J]. International Biodeterioration & Biodegradation, 2010, 64(4): 324-330. |
| [46] |
POOJA T, VASUDHA A, ANITA P, et al. Biodegradation of bisphenol A using psychrotolerant bacterial strain Pseudomonas palleroniana GBPI_508[J]. Archives of Microbiology, 2022, 204(5): 272-284. DOI:10.1007/s00203-022-02885-y |
| [47] |
TIAN K, YU Y, QIU Q, et al. Mechanisms of BPA degradation and toxicity resistance in Rhodococcus equi[J]. Microorganisms, 2023, 11(1): 67-83. |
| [48] |
NORDIN K, UNELL M, JANSSON J K. Novel 4-chlorophenol degradation gene cluster and degradation route via hydroxyquinol in Arthrobacter chlorophenolicus A6[J]. Applied and Environmental Microbiology, 2005, 71(11): 6538-6544. DOI:10.1128/AEM.71.11.6538-6544.2005 |
| [49] |
BAE H S, LEE J M, LEE S T. Biodegradation of 4-chlorophenol via a hydroquinone pathway by Arthrobacter ureafaciens CPR706[J]. FEMS Microbiology Letters, 1996, 145(1): 125-129. DOI:10.1111/j.1574-6968.1996.tb08566.x |
| [50] |
KONOVALOVA E, SOLYANIKOVA I, GOLOVLEVA L. Degradation of 4-chlorophenol by the strain Rhodococcus opacus 6a[J]. Microbiology, 2009, 78(6): 805-807. DOI:10.1134/S0026261709060204 |
| [51] |
ZHENG J, ZHANG Z, AN J, et al. Adaptive laboratory evolution of Rhodococcus rhodochrous DSM6263 for chlorophenol degradation under hypersaline condition[J]. Microbial Cell Factories, 2023, 22(1): 220-239. DOI:10.1186/s12934-023-02227-7 |
| [52] |
MATUS V, SANCHEZ M, MARTINEZ M, et al. Efficient degradation of 2, 4, 6-trichlorophenol requires a set of catabolic genes related to tcp genes from Ralstonia eutropha JMP134(pJP4)[J]. Applied and Environmental Microbiology, 2003, 69(12): 7108-7115. DOI:10.1128/AEM.69.12.7108-7115.2003 |
| [53] |
LOUIE T M, WEBSTER C M, XUN L. Genetic and biochemical characterization of a 2, 4, 6-trichlorophenol degradation pathway in Ralstonia eutropha JMP134[J]. Journal of Bacteriology, 2002, 184(13): 3492-3500. DOI:10.1128/JB.184.13.3492-3500.2002 |
| [54] |
WEBB B N, BALLINGER J W, KIM E, et al. Characterization of chlorophenol 4-monooxygenase(TftD) and NADH: FAD oxidoreductase(TftC) of Burkholderia cepacia AC1100[J]. Journal of Biological Chemistry, 2010, 285(3): 2014-2027. DOI:10.1074/jbc.M109.056135 |
| [55] |
OLANIRAN A O, SINGH L, KUMAR A, et al. Aerobic degradation of 2, 4-dichlorophenoxyacetic acid and other chlorophenols by Pseudomonas strains indigenous to contaminated soil in South Africa: Growth kinetics and degradation pathway[J]. Applied Biochemistry and Microbiology, 2017, 53(2): 209-216. DOI:10.1134/S0003683817020120 |
| [56] |
LI D Y, EBERSPCHER J, WAGNER B, et al. Degradation of 2, 4, 6-trichlorophenol by Azotobacter sp. strain GP1[J]. Applied and Environmental Microbiology, 1991, 57(1): 1920-1928. |
| [57] |
KAO C M, LIU J K, CHEN Y L, et al. Factors affecting the biodegradation of PCP by Pseudomonas mendocina NSYSU[J]. Journal of Hazardous Materials, 2005, 124(1): 68-73. |
| [58] |
ITO K, TAKAGI K, MATSUSHIMA Y, et al. Identification of the novel hcbB operon catalyzing the dechlorination of pentachlorophenol in the Gram-positive bacterium Nocardioides sp. strain PD653[J]. Journal of Pesticide Science, 2018, 43(2): 124-131. DOI:10.1584/jpestics.D17-089 |
| [59] |
OHTSUBO Y, MIYAUCHI K, KANDA K, et al. PcpA, which is involved in the degradation of pentachlorophenol in Sphingomonas chlorophenolica ATCC39723, is a novel type of ring-cleavage dioxygenase[J]. FEBS Letters, 1999, 459(3): 395-398. DOI:10.1016/S0014-5793(99)01305-8 |
| [60] |
TUAN N N, HSIEH H C, LIN Y W, et al. Analysis of bacterial degradation pathways for long-chain alkylphenols involving phenol hydroxylase, alkylphenol monooxygenase and catechol dioxygenase genes[J]. Bioresource Technology, 2011, 102(5): 4232-4240. DOI:10.1016/j.biortech.2010.12.067 |
| [61] |
BAI N, ABUDUAINI R, WANG S, et al. Nonylphenol biodegradation characterizations and bacterial composition analysis of an effective consortium NP-M2[J]. Environmental Pollution, 2017, 220(Jan.): 95-104. |
| [62] |
WANG H, DENG W, SHEN M H, et al. A laccase Gl-LAC-4 purified from white-rot fungus Ganoderma lucidum had a strong ability to degrade and detoxify the alkylphenol pollutants 4-n-octylphenol and 2-phenylphenol[J]. Journal of Hazardous Materials, 2021, 408(Apr.): 124775.1-16. |
| [63] |
WANG J, ZHANG L, HE Y, et al. Biodegradation of phenolic pollutants and bioaugmentation strategies: A review of current knowledge and future perspectives[J]. Journal of Hazardous Materials, 2024, 469: 133906. DOI:10.1016/j.jhazmat.2024.133906 |
| [64] |
ZHOU N A, KJELDAL H, GOUGH H, et al. Identification of putative genes involved in bisphenol A degradation using differential protein abundance analysis of Sphingobium sp. BiD32[J]. Environmental Science & Technology, 2015, 49(20): 12232-12241. |
| [65] |
ROH H, SUBRAMANYA N, ZHAO F, et al. Biodegradation potential of wastewater micropollutants by ammonia-oxidizing bacteria[J]. Chemosphere, 2009, 77(8): 1084-1089. DOI:10.1016/j.chemosphere.2009.08.049 |
| [66] |
TELKE A A, KALYANI D C, JADHAV U U, et al. Pur-ification and characterization of an extracellular laccase from a Pseudomonas sp. LBC1 and its application for the removal of bisphenol A[J]. Journal of Molecular Catalysis B: Enzymatic, 2009, 61(3): 252-260. |
| [67] |
GUO E, ZHAO L, LI Z, et al. Biodegradation of bisphenol A by a Pichia pastoris whole-cell biocatalyst with overexpression of laccase from Bacillus pumilus and investigation of its potential degradation pathways[J]. Journal of Hazardous Materials, 2024, 474: 134779. DOI:10.1016/j.jhazmat.2024.134779 |
| [68] |
WANG X W, CHEN J Q, JI R, et al. Degradation of bisphenol S by a bacterial consortium enriched from River Sediments[J]. Bulletin of Environmental Contamination and Toxicology, 2019, 103(4): 100314. |
| [69] |
YIN R, ZHANG R, WANG B J, et al. Biotransformation of bisphenol F by white-rot fungus Phanerochaete sordida YK-624 under non-ligninolytic condition[J]. Applied Microbiology and Biotechnology, 2022, 106(18): 6277-6287. DOI:10.1007/s00253-022-12133-4 |
| [70] |
WANG J Q, YIN R, ZHANG X, et al. Transcriptomic analysis reveals ligninolytic enzymes of white-rot fungus Phanerochaete sordida YK-624 participating in bisphenol F biodegradation under ligninolytic conditions[J]. Environmental Science and Pollution Research International, 2021, 28(44): 62390-62397. DOI:10.1007/s11356-021-15012-z |
| [71] |
FARRELL A, QUILTY B. Degradation of mono-chlorophenols by a mixed microbial community via a meta-cleavage path-way[J]. Biodegradation, 1999, 10(5): 353-362. DOI:10.1023/A:1008323811433 |
| [72] |
DORN E, KNACKMUSS H. Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol 1, 2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad[J]. Biochemical Journal, 1978, 174(1): 73-84. DOI:10.1042/bj1740073 |
| [73] |
XUN L, WEBSTER C M. A monooxygenase catalyzes sequential dechlorinations of 2, 4, 6-trichlorophenol by oxidative and hydrolytic reactions[J]. Journal of Biological Chemistry, 2004, 279(8): 6696-6700. DOI:10.1074/jbc.M312072200 |
| [74] |
UOTILA J, SALKINOJA-SALONEN M, APAJALAHTI J. Dechlorination of pentachlorophenol by membrane bound enzymes of Rhodococcus chlorophenolicus PCP-I[J]. Biodegradation, 1991, 2(1): 25-31. DOI:10.1007/BF00122422 |
| [75] |
UOTILA J, KITUNEN V, SAASTAMOINEN T, et al. Characterization of aromatic dehalogenases of Mycobacterium fortuitum CG-2[J]. Journal of Bacteriology, 1992, 174(17): 5669-5675. DOI:10.1128/jb.174.17.5669-5675.1992 |
| [76] |
DUAN X, WANG X, XIE J, et al. Acidogenic bacteria assisted biodegradation of nonylphenol in waste activated sludge during anaerobic fermentation for short-chain fatty acids production[J]. Bioresource Technology, 2018, 268: 692-699. DOI:10.1016/j.biortech.2018.08.053 |
| [77] |
WANG Z, YANG Y, DAI Y, et al. Anaerobic biodegradation of nonylphenol in river sediment under nitrate-or sulfate-reducing conditions and associated bacterial community[J]. Journal of Hazardous Materials, 2015, 286: 306-314. DOI:10.1016/j.jhazmat.2014.12.057 |
| [78] |
YANG Y Y, WANG Z, HE T, et al. Sediment bacterial communities associated with anaerobic biodegradation of bisphenol A[J]. Microbial Ecology, 2015, 70(1): 97-104. DOI:10.1007/s00248-014-0551-x |
| [79] |
LI Z, SUZUKI D, ZHANG C, et al. Involvement of Dehalobacter strains in the anaerobic dechlorination of 2, 4, 6-trichl-orophenol[J]. Journal of Bioscience and Bioengineering, 2013, 116(5): 602-609. DOI:10.1016/j.jbiosc.2013.05.009 |
| [80] |
ANNIE G, REJEAN B, FRANCOIS L, et al. Occurrence and expression of crdA and cprA5 encoding chloroaromatic reductive dehalogenases in Desulfitobacterium strains[J]. Canadian Journal of Microbiology, 2006, 52(1): 47-55. DOI:10.1139/w05-111 |
| [81] |
VILLEMUR R. The pentachlorophenol-dehalogenating Desulfitobacterium hafniense strain PCP-1[J]. Philosophical Transactions of the Royal Society of London, Series B, Biological Sciences, 2013, 368(1616): 1-18. |
| [82] |
ZHAO Y, JI J, WU Y, et al. Nonylphenol and its derivatives: Environmental distribution, treatment strategy, management and future perspectives[J]. Chemosphere, 2024, 352: 141377. DOI:10.1016/j.chemosphere.2024.141377 |
| [83] |
CHANG B, YU C, YUAN S. Degradation of nonylphenol by anaerobic microorganisms from river sediment[J]. Chemosphere, 2004, 55(4): 493-500. DOI:10.1016/j.chemosphere.2004.01.004 |
| [84] |
CHANG B, CHIANG F, YUAN S. Anaerobic degradation of nonylphenol in sludge[J]. Chemosphere, 2005, 59(10): 1415-1420. DOI:10.1016/j.chemosphere.2004.12.055 |
| [85] |
GONZALEZ-GIL L, CARBALLA M, LEMA J M. Cometabolic enzymatic transformation of organic micropollutants under methanogenic conditions[J]. Environmental Science & Technology, 2017, 51(5): 2963-2971. |
| [86] |
KANG J H, KONDO F. Bisphenol a degradation by bacteria isolated from river water[J]. Archives of Environmental Contamination and Toxicology, 2002, 43(3): 265-269. DOI:10.1007/s00244-002-1209-0 |
| [87] |
VOORDECKERS J W, FENNELL D E, JONES K, et al. Anaerobic biotransformation of tetrabromobisphenol A, tetrachlorobisphenol A, and bisphenol A in estuarine sediments[J]. Environmental Science & Technology, 2002, 36(4): 696-701. |
| [88] |
YADAV S, KUMAR S, HARITASH A K. A comprehensive review of chlorophenols: Fate, toxicology and its treatment[J]. Journal of Environmental Management, 2023, 342: 118254. DOI:10.1016/j.jenvman.2023.118254 |
| [89] |
OLANIRAN A O, IGBINOSA E O. Chlorophenols and other related derivatives of environmental concern: Properties, distribution and microbial degradation processes[J]. Chemosphere, 2011, 83(10): 1297-1306. DOI:10.1016/j.chemosphere.2011.04.009 |
| [90] |
El-SAYED W S. Characterization of a highly enriched microbial consortium reductively dechlorinating 2, 3-dichlorophenol and 2, 4, 6-trichlorophenol and the corresponding cprA genes from river sediment[J]. Polish Journal of Microbiology, 2016, 65(3): 341-352. DOI:10.5604/17331331.1215613 |
| [91] |
SONG J, CHEN L, CHEN H, et al. Characterization and high-throughput sequencing of a trichlorophenol-dechlorinating microbial community acclimated from sewage sludge[J]. Journal of Cleaner Production, 2018, 197: 306-313. DOI:10.1016/j.jclepro.2018.06.061 |
 2024, Vol. 16
2024, Vol. 16 

