孔雀石绿(malachite green)是一种人工合成的三苯甲烷类化合物(图 1),曾在水产养殖业中作为杀菌剂和杀虫剂被广泛使用。20世纪90年代以来,国内外学者陆续发现孔雀石绿具有广泛毒性,可以引起代谢紊乱[1]、DNA损伤[1],甚至诱发促进肿瘤生长[2-4],危害人类健康,因此,我国在《无公害食品渔用药物使用准则》(NY 5071—2002)中将孔雀石绿列为禁用药物。但由于廉价替代品的缺乏,孔雀石绿在我国多个省份的水产养殖业依然屡禁不止[5-10],且在中国检测鳗鱼体内的孔雀石绿最高平均值为30.75 μg/kg,中国海洋鱼类检测出的最低报告平均值为0.000 25 μg/kg,根据食品法典委员会限值(MRLP=2 μg/kg),中国(4.157 μg/kg)的鱼类样本中总孔雀石绿的平均质量分数高于允许水平[11]。除此之外,孔雀石绿也是一种染料,曾在或依然在印染、印刷、化妆品等行业中被广泛使用[12],还可以作为工业材料杀菌剂、细胞染色剂、酸碱指示剂等使用。
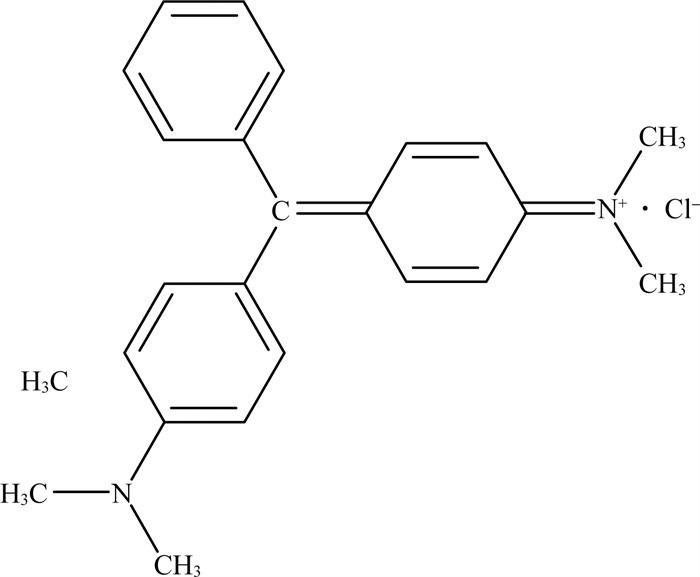
|
图 1 孔雀石绿盐酸盐形式(CAS:569-64-2)结构示意 |
由于孔雀石绿在水产养殖业中依然屡禁不止,且在印染等行业亦被广泛使用,这使其在环境中仍然大量残留,并能通过食物链进一步富集并严重危害人类健康。因此研究高效、低成本、安全降解孔雀石绿的方式具有重要科学与应用意义。生物降解相较于传统理化降解是一种更加高效、廉价且安全的方式。国内外已有许多学者相继开展了微生物降解孔雀石绿的研究。然而,目前尚没有有关微生物降解孔雀石绿的详尽文献综述。本研究从可降解菌种、降解条件、降解途径、降解酶系、固定化进展、组学进展6个方面,系统总结了微生物2大主要类群(细菌与真菌)降解孔雀石绿的研究进展。以期为进一步开展孔雀石绿微生物降解的机理研究与技术应用提供科学论据。
1 细菌降解孔雀石绿研究进展 1.1 可降解菌种可降解孔雀石绿细菌菌种信息见表 1。由表 1可见,可降解孔雀石绿的细菌共30种,分属19个属。其中,11个属(占比57.89%)归于变形菌门(Proteobacteria),5个属(占比26.32%)归于厚壁菌门(Firmicutes),其他3个属(占比15.79%)归于放线菌门(Actinobacteria)或无明确分类。
| 表 1 可降解孔雀石绿细菌菌种① |
大部分细菌或其提纯酶均可在24 h内实现超过90%孔雀石绿的降解,甚至达到完全降解,如产气克雷伯氏菌S27(Klebsiella aerogenes S27)可在8 h内将质量浓度为100 mg/L的孔雀石绿完全降解。某些细菌可在高浓度孔雀石绿下存活,如假单胞菌DY1(Pseudomonas sp. DY1)、阿氏肠杆菌XJUHX-4TM(Enterobacter asburiae XJUHX-4TM)、塞氏柠檬酸杆菌RI11(Citrobacter sedlakii RI11)和假单胞菌YB2(Pseudomonas sp. YB2)等,它们均可降解质量浓度为1 000 mg/L甚至更高浓度的孔雀石绿。
1.2 最适降解条件 1.2.1 温度温度可以影响细菌或酶对孔雀石绿的降解效率。不同细菌菌种或酶降解孔雀石绿的最适温度见表 2。由表 2可见,大部分细菌或酶的最适降解温度为25~40 ℃。仅有耐辐射奇球菌R1(Deinococcus radiodurans R1)能够适应较广的温度范围,温度最高可达50 ℃[22]。
| 表 2 不同细菌菌种或酶降解孔雀石绿最适温度 |
培养环境的pH值可以影响细菌繁殖和生长,从而影响细菌或酶对孔雀石绿的降解效率。不同细菌菌种降解孔雀石绿的最适pH值见表 3。由表 3可见,细菌或酶的最适降解pH值均在偏中性—弱碱性范围。
| 表 3 不同细菌菌种降解孔雀石绿最适pH值 |
培养基中的碳源、氮源、金属离子等物质会影响细菌对孔雀石绿的降解效果。
Deng等[14]研究了11种不同碳源下蜡样芽孢杆菌DC11(Bacillus cereus DC11)对孔雀石绿降解效率的差异,发现额外添加碳源会使孔雀石绿降解率提高10%~40%,其中酵母提取物、葡萄糖和麦芽糖作为碳源时降解效果最好,降解率高达100%。Vignesh等[44]也得到了相似结论,即添加1%葡萄糖和酵母提取物可提高孔雀石绿降解速率90%。Wu等[18]研究也表明,在培养基中加入酵母提取物,会提高耳炎假单胞菌WL-13(Pse-udomonas otitidis WL-13)的降解效率。此外,李刚等[37]将淀粉作为培养基碳源,也能提高肠杆菌CV-b(Enterobacter sp. CV-b)对孔雀石绿降解效率。除碳源影响外,氮源对细菌生长与对污染物降解也具有显著影响。Mukherjee等[25]发现,当蔗糖作为碳源、牛肉提取物作为氮源,且碳氮比(C/N)为5 ∶ 1时,阿氏肠杆菌XJUHX-4TM(Enterobacter asburiae XJUHX-4TM)对孔雀石绿降解率可达98%。李刚等[37]和Sutar等[43]均发现蛋白胨作为氮源,可以促进细菌降解孔雀石绿。
研究表明,培养基中加入1 mmol/L镁离子和锰离子可显著促进假单胞菌DY1(Pseudomonas sp.DY1)对孔雀石绿降解效率[21];Li等[42]也发现添加镁离子、锰离子和铜离子可促进肺炎克雷伯菌WA-1(Klebsiella pneumoniae WA-1)降解效率;刘单单等[40]也提出了相似结论,即低浓度铁离子、锰离子、铜离子和铅离子或可提高肠杆菌B-20(Enterobacter sp. B-20)降解孔雀石绿效率。但是,某些金属离子也会抑制细菌降解孔雀石绿,如铜离子对肠杆菌CV-b(Enterobacter sp. CV-b)降解孔雀石绿有显著抑制作用[37],铬离子会抑制洋葱伯克霍尔德菌C09G(Burkholderia cepacia C09G)降解孔雀石绿[35],钙离子、铁离子、铅离子和锌离子会抑制肺炎克雷伯菌WA-1(Klebsiella pneumoniae WA-1)的降解效率[42]。
有研究表明,在培养基中添加0.075%的微生物表面活性剂或可显著提高塞氏柠檬酸杆菌RI11(Citrobacter sedlakii RI11)对孔雀石绿降解效率,而添加合成的表面活性剂则具有效果相反[26]。
1.2.4 孔雀石绿初始浓度Ayed等[16]研究了少动鞘氨醇单胞菌(Sphingomonas paucimobilis)对不同初始浓度孔雀石绿的降解能力,发现孔雀石绿质量浓度为2.5~50 mg/L时,随着浓度增加,细菌在24 h内对孔雀石绿的降解率由100%降低到80%,这可能是由于高浓度孔雀石绿对细菌有更强毒害所致。肺炎克雷伯菌WA-1(Klebsiella pneumoniae WA-1)对质量浓度为1~80 mg/L的孔雀石绿[42],苍白杆菌JN214485(Ochrobactrum sp. JN214485)对质量浓度为50~200 mg/L的孔雀石绿[24]以及嗜水气单胞菌(Pseudomonas veronii)对质量浓度为20~200 mg/L的孔雀石绿[46]的降解动力学亦有类似情况。此外,Lv等[22]、Vignesh等[44]和Mnif等[26]研究表明,耐辐射奇异球菌R1(Deinococcus radiodurans R1)、天蓝色链霉菌S20(Strepto-myces chrestomyceticus S20)和塞氏柠檬酸杆菌RI11(Citrobacter sedlakii RI11)分别在更广浓度范围内,即50~500,100~500,50~1 000 mg/L,对孔雀石绿降解效率也与初始浓度呈现负相关。
与之相反,Chen等[17]研究发现居肺潘多拉菌YC32(Pandoraea pulmonicola YC32)在质量浓度为10~100 mg/L时对孔雀石绿的降解率随着质量浓度升高而增大,且在孔雀石绿质量浓度为100 mg/L时,其降解率与浓度符合林尤韦伯-伯克(Line-weaver-Burk)方程。同样,枯草芽孢杆菌cjp3(Bacillus subtilis cjp3)在质量浓度为25~100 mg/L时,降解速度与孔雀石绿浓度呈现正相关[41]。
1.2.5 其他因素氧浓度也会影响细菌对污染物的降解效率,特别是通过摇晃培养器皿能够增大液体培养基与空气接触面积从而增大培养基的氧浓度。蜡样芽孢杆菌DC11(Bacillus cereus DC11)在低氧/厌氧条件下对孔雀石绿的降解效果较好氧条件下好[14]。而塞氏柠檬酸杆菌RI11(Citrobacter sedlakii RI11)在静态培养和摇晃培养2种条件下虽然都能够降解孔雀石绿,但摇晃培养降解率(>80%)明显高于静态培养降解率(14%~40%)[26]。同样地,海洋发光细菌MS(Photobacterium leiognathi MS)在摇晃培养降解率(92.04%)亦高于静态培养降解率(83.73%)[43]。此外,假单胞菌DY1(Pseudomonas sp. DY1)可降解较高质量浓度(100~1 000 mg/L)的孔雀石绿,但在摇晃培养条件下具有较高的降解率(90.3%~97.2%),而在静态培养时降解率较低(78.9%~84.3%)[21]。对众多不同文献中采用的培养条件(摇晃培养或静态培养)进行总结,其中有21篇文献研究(63.64%)采取摇晃培养,2篇(6.06%)采取静态培养,5篇(15.15%)并未提及摇晃或静态的降解条件。由此可以初步看出,可降解孔雀石绿的细菌多数为好氧细菌。
接种量也会影响细菌降解孔雀石绿效率。例如,少动鞘氨醇单胞菌(Sphingomonas paucimobilis)[16]和嗜水气单胞菌(Pseudomonas veronii)[46],随着其接种量的增大,孔雀石绿降解率呈逐步升高趋势。又如,肺炎克雷伯菌WA-1(Klebsiella pneumoniae WA-1)在降解质量浓度为40 mg/L的孔雀石绿时,接种量为0.4 g/L时降解率比接种量为0.025 g/L时增加了15.4倍,而其接种量对低质量浓度的孔雀石绿降解效率几乎没有显著影响[42]。
1.3 降解途径细菌降解孔雀石绿路径已被研究得较为详尽透彻,具体见图 2[14, 15, 17, 20-22, 24, 25, 28, 29, 31, 34, 39, 43, 46]。降解路径可大致总结为N-去甲基化和中心碳原子羟基化2种。
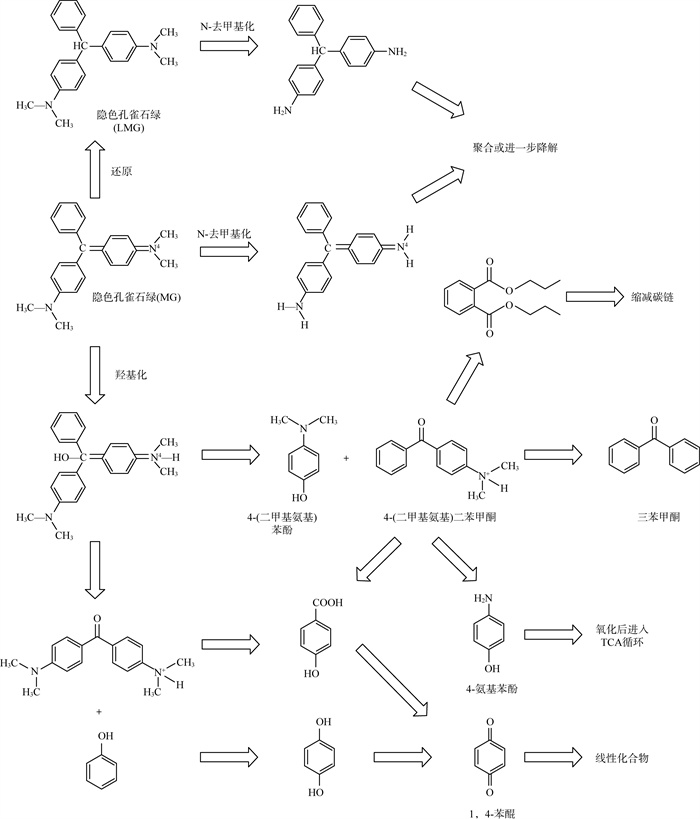
|
图 2 细菌降解孔雀石绿途径路线 |
孔雀石绿可以通过N-去甲基化脱去4个甲基,也可以先还原为隐色孔雀石绿后进行N-去甲基化脱去4个甲基,后进行聚合或进一步降解。
孔雀石绿中心碳原子羟基化后分解为4-(二甲基氨基)苯酚和4-(二甲基氨基)二苯甲酮,后者能进一步降解为二苯甲酮或4-氨基苯酚氧化后进入三羧酸循环,或者进行开环和缩短碳链。中心碳原子羟基化后,或通过去苯基化和氧化反应生成1,4-苯醌,最后经过开环反应和氧化反应再分解为一系列线性化合物。
1.4 降解酶参与降解孔雀石绿的酶包括漆酶、三苯基甲烷还原酶、锰过氧化物酶、孔雀石绿还原酶、烟酰胺腺嘌呤二核苷酸-二氯苯酚还原酶(NADH-DCIP还原酶)、氨基比林N-脱甲基酶、二氯酚视黄醇还原酶和酪氨酸酶等,具体见表 4。此外,Qu等[47]基于宏基因组测序发现编码孔雀石绿降解酶基因中,过氧化物酶基因最多(93.54%),漆酶基因和P450酶基因较少(分别3.40%和3.06%)。
| 表 4 参与降解孔雀石绿的细菌降解酶 |
在实际应用中,细菌菌体的固定化可提高其对孔雀石绿降解效率,并可进一步应用于含孔雀石绿染料废水的处理。Vijayalakshmidevi等[24]利用无菌海藻酸钙珠将苍白杆菌JN214485(Ochrobactrum sp. JN214485)固定化,当微珠负载量为20 g/L时,处理48 h后,降解率高达80%。Gan等[29]将洋葱伯克霍尔德菌Burkholderia cepacia固定于桉树叶片上,制备成了可以同时降解孔雀石绿和重金属铬离子(Cr6+)的复合材料,可同时去除94.8%的孔雀石绿和71.9%的重金属铬;Li等[30]将洋葱伯克霍尔德菌Burkholderia cepacia固定于农业生物废料——甘蔗渣上,可去除98.5%孔雀石绿,比单纯细菌处理去除率高约10%;且这2种材料对于孔雀石绿去除均符合拟二级吸附动力学模型。
除菌体固定化外,Sinirlioglu等[23]将腐败希瓦氏菌(Shewanella putrefaciens)中提纯所得漆酶进行固定化,制成交联酶聚合体(cross-linked enzyme aggregates,CLEA),应用于工业含染料废水处理,可在24 h内去除90%初始质量浓度为50 mg/L的孔雀石绿废水。Qiao等[41]将枯草芽孢杆菌cjp3(Bacillus subtilis cjp3)提纯所得漆酶固定于磁性氧化石墨烯纳米材料上制成磁性复合材料(MGO-漆酶),其较游离漆酶具有更强的酸碱稳定性、热稳定性和耐受抑制剂或金属离子能力。
1.6 组学研究当前通过组学技术研究细菌降解孔雀石绿的分子机制的文献报道较少。Wang等[27]和Qu等[39]分别对食油假单胞菌MGY01(Pseudomonas oleovorans MGY01)和黏着杆菌HMG1(Tenacibaculum sp. HMG1)进行了全基因组测序与分析。此外,还有Qu等[47]利用红树林沉积物宏基因组测序技术挖掘孔雀石绿降解基因,并评估其相应酶的潜力,共发现44个与孔雀石绿降解相关基因,推测可能来源于30个细菌属,大部分属于变形菌门和拟杆菌门(Bacteroidetes)。这与本研究所得结论一致,即可降解孔雀石绿细菌在进化分类上大部分属于变形菌门。
2 真菌降解孔雀石绿研究进展 2.1 可降解菌种可降解孔雀石绿的真菌菌种见表 5。可降解孔雀石绿真菌共有55种,归于27个属;其中,来自担子菌门(Basidiomycota)-伞菌纲(Agarico-mycetes)-多孔菌目(Polyporales)的有13个属(约48.15%),而来自担子菌门其他目的有6个属(约22.22%);此外,来自子囊菌门(Ascomycota)的有7个属(约25.93%),而来自毛霉菌门(Muco-romycota)的仅有1个属(约3.70%)。
| 表 5 可降解孔雀石绿的真菌菌种① |
大部分真菌或其提纯酶降解孔雀石绿所需的时间较细菌更长,其中有53种真菌或其提纯酶的降解时间超过1d,整体上降解率为2.1%~100%。可以将孔雀石绿完全降解的真菌菌种多于细菌菌种,共有11种,其中Fomes sclerodermeus BAFC 2752菌、硬毛栓孔菌BAFC 463(Trametes trogii BAFC 463)、灵芝rckk-02(Ganoderma sp. rckk-02)、黑孔菌HYB07(Cerrena sp. HYB07)和云芝栓孔菌(Trametes versicolor)都是提纯漆酶实现了孔雀石绿的完全降解。
此外,与细菌降解比较发现,真菌对于高浓度孔雀石绿的耐受能力相对较差,有文献报道真菌最高可降解质量浓度为1 000 mg/L的孔雀石绿,且降解率不足20%[48],而细菌最高可降解质量浓度为1 500 mg/L的孔雀石绿[28]。
2.2 最适降解条件 2.2.1 温度温度可以影响真菌生长繁殖,也能影响降解酶活性和稳定性,进而影响孔雀石绿的降解效率。不同真菌菌种或其提纯酶降解孔雀石绿最适温度见 表 6。由表 6可见,真菌降解孔雀石绿最适温度接近室温,这与细菌相似;而真菌提纯酶降解孔雀石绿最适温度则相对较高,甚至达到了70 ℃,可见真菌提纯降解酶对温度的耐受程度和稳定性更强。
| 表 6 不同真菌菌种或其提纯酶降解孔雀石绿最适温度 |
环境pH值可以影响真菌生长繁殖,还能通过影响细胞膜运输和降解酶活性。不同真菌菌种降解孔雀石绿最适pH值见表 7。由表 7可见,与细菌最适pH值不同,真菌或其提纯酶最适pH值均偏中性或酸性,甚至可以耐受强酸性环境。例如,弯孢拟蜡孔菌ATCC90467(Ceriporiopsis subvermispora ATCC90467)提纯漆酶最适pH值=2[76]。
| 表 7 不同真菌菌种降解孔雀石绿最适pH值① |
培养基的碳源和氮源是真菌生长繁殖的营养来源,直接影响真菌生长和产酶速度。例如,对于露湿漆斑菌IM6482(Myrothecium roridum IM6482),碳源种类只在降解前8 h有影响,而处理24 h后,不同碳源组别降解率无明显差异,均接近100%[70]。又如,谷氨酸和蛋白胨是硬毛栓孔菌(Trametes trogii)、长绒毛栓菌(Trametes villosa)和云芝(Coriolus versicolor var. antarcticus)生产漆酶和锰过氧化物酶的最佳氮源[60]。葡萄糖作为碳源、尿素作为氮源或可增强黄孢原毛平革菌(Phanerochaete chrysosporium)对孔雀石绿降解[55];蔗糖和硝酸钠分别是黄曲霉菌(Aspergillus flavus)降解时有效碳源和氮源[48];葡萄糖和酵母浸粉分别为裂褶菌cfcc7252(Schizophyllum commune cfcc7252)降解的最佳碳源和氮源[92]。此外,Marcharchand等[91]研究了培养基浓度对棘孢木霉菌(Trichoderma asperellum)降解孔雀石绿的影响,发现低营养水平会影响真菌生长,但对孔雀石绿降解无显著影响。Morales-Álvarez等[88]发现较低浓度的碳源和氮源(2.5 g/L葡萄糖和0.05 g/L氯化铵)有利于灵芝(Ganoderma lucidum)降解孔雀石绿。
金属离子也对真菌降解孔雀石绿产生一定影响。研究表明,锰离子(Mn2+)、镁离子(Mg2+)、锌离子(Zn2+)、钴离子(Co2+)、钠离子(Na+)(100 mmol/L)对硬毛栓孔菌S0301(Trametes trogii Berk S0301)提纯漆酶降解孔雀石绿产生轻微抑制(< 10%)[69]。添加浓度为2.0 mmol/L的Cu2+可以通过促进栓孔菌LAC-01(Trametes sp. LAC-01)产生漆酶,从而促进孔雀石绿降解[73]。Ca2+可以通过增加线粒体膜电位和增强细胞色素C氧化酶的活性促进黑曲霉菌(Aspergillus niger)对孔雀石绿降解[82]。
向培养基中添加其他物质也可影响真菌或提纯降解酶对孔雀石绿降解效果。如添加1-羟基苯并三唑[51, 58, 65, 74]、天然酚类化合物[57, 63]、乙酰丙酮[75]、紫尿酸和乙酰丁香酮[85]可以促进漆酶对孔雀石绿降解,而单宁酸[58]则有相反作用。相似地,全氟辛烷磺酸[86]可以通过抑制锰过氧化物酶和木质素过氧化物酶的活性进而抑制其对孔雀石绿降解。
2.2.4 孔雀石绿初始浓度Chen等[71]比较了革孔菌1c3(Coriolopsis sp. 1c3)对不同浓度孔雀石绿的降解情况,发现其对质量浓度为50 mg/L孔雀石绿在5 d内可降解98%;质量浓度为100 mg/L孔雀石绿处理9 d后可降解52%;而质量浓度为200 mg/L时,最终降解率仅为6%。黄曲霉菌(Aspergillus flavus)对于质量浓度为100~1 000 mg/L的孔雀石绿降解实验表明,浓度越高,降解率越低[48]。相似还有毛色二孢菌(Lasiodiplodia sp.)对质量浓度为50~500 mg/L的孔雀石绿降解率随着溶液浓度升高而降低(降解率由96.9%降低到89.18%)[100]。
Sharma等[74]比较了灵芝rckk-02(Ganoderma sp. rckk-02)提纯漆酶对不同浓度孔雀石绿的降解情况,漆酶对初始浓度为30 U/mL(代表每毫升药品中含有多少单位药物),100 mg/L和200 mg/L的孔雀石绿在16~20 h内可完全降解;对质量浓度为300 mg/L的孔雀石绿处理24 h降解率为72%;对质量浓度为400 mg/L的孔雀石绿处理28 h后降解率为62%,对质量浓度为500 mg/L的孔雀石绿处理32 h后降解率为55%。而黄孢原毛平革菌(Phanerochaete chrysosporium)提纯漆酶对较高浓度孔雀石绿降解率则较低,在溶液质量浓度<12 mg/L时,可在24 h内降解94%,但对于质量浓度为25 mg/L的孔雀石绿,则几乎没有降解能力[102]。
2.2.5 其他因素真菌对氧含量需求有所差异,静态培养与摇晃培养应依据真菌的特性而选择。例如,黄曲霉菌(Aspergillus flavus)[48]和黄孢原毛平革菌(Phanerochaete chrysosporium)[67]更适宜摇晃培养,黑曲霉菌(Aspergillus niger)[67]和革孔菌1c3(Coriolopsis sp. 1c3)[71]则更适宜静态培养,而氧含量对露湿漆斑菌IM6482(Myrothecium roridum IM 6482)[70]和裂褶菌cfcc7252(Schizophyllum commune cfcc7252)[92]降解孔雀石绿几乎没有影响。对不同文献中采用的培养条件(摇晃培养或静态培养)进行总结,其中有12篇文献(21.82%)采取摇晃培养,3篇(5.45%)采取静态培养,35篇(63.64%)并未提及摇晃或静态的降解条件。虽然多数文献并未提及,但是因真菌为真核生物,新陈代谢方式多数为需氧型,可初步推断,摇晃培养有助于真菌降解孔雀石绿。
接种量也可以影响孔雀石绿降解效率。如增加革孔菌1c3(Coriolopsis sp. 1c3)接种量(1~8 g)可提高孔雀石绿降解率,当接种量为8 g时,降解率为70%[71]。Morales-Álvarez等[88]也发现菌丝接种量越大,对孔雀石绿的降解率越高。
2.3 降解途径真菌对孔雀石绿的降解途径较为明晰,将已知所有降解途径进行汇总,具体见图 3 [48,59,63,78-79,89- 90,93,99,102]。降解路径可大致总结为N-去甲基化和中心碳原子羟基化2种。
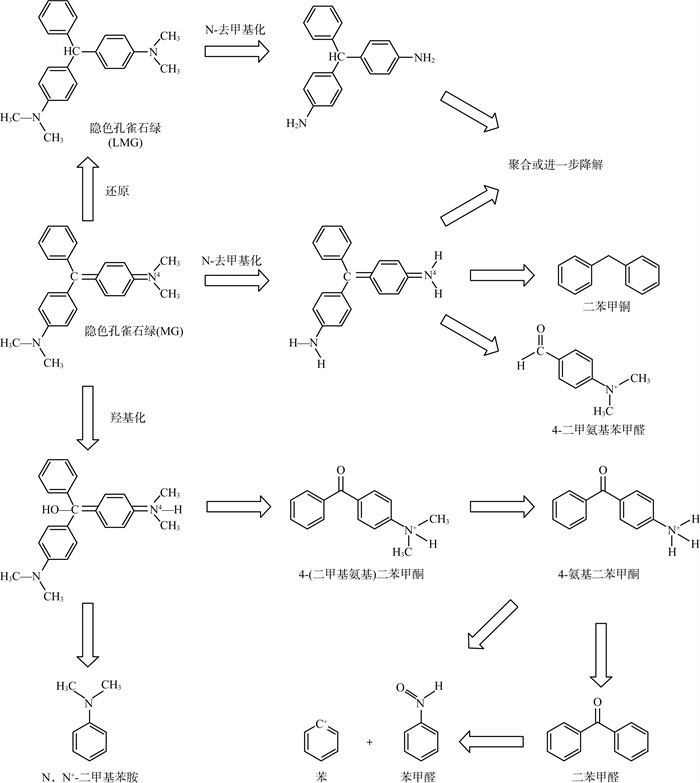
|
图 3 真菌对孔雀石绿的降解途径路线 |
孔雀石绿在真菌或提纯酶的作用下还原成隐色孔雀石绿后经历4个连续N-去甲基化步骤,也可直接N-去甲基化,进一步可降解成二苯甲烷和4-二甲氨基苯甲醛。
孔雀石绿经历中心碳原子的羟基化,随后分解得到4-(二甲基氨基)二苯甲酮和N,N′-二甲基苯胺,前者可进一步N-去甲基化生成二苯甲酮,然后进一步降解得到苯和苯甲醛。
综上,真菌最终降解产物均是含苯环的有机化合物,而细菌则可将含苯环中间产物经过开环和氧化反应再分解为一系列线性化合物,因此真菌对孔雀石绿的降解程度不及细菌。
2.4 降解酶参与真菌降解孔雀石绿的酶包括漆酶、锰过氧化物酶、木质素过氧化物酶、NADH-DCIP还原酶、三苯基甲烷还原酶和酪氨酸酶等,具体见表 8。
| 表 8 参与降解孔雀石绿的真菌降解酶 |
此外,Jasinska等[70]利用酶抑制实验初步证实露湿漆斑菌IM 6482(Myrothecium roridum IM 6482)降解孔雀石绿没有过氧化物酶和CYP450酶的参与。
2.5 固定化真菌或其提纯酶通过固定化制备成复合体较游离菌体或游离酶有更好的降解率、稳定性和重复使用性等,因此,有学者将真菌固定于选定材料上,以应用于含染料废水的处理。
Yang等[68]制备了负载血红密孔菌(Pycnoporus sanguineus)的磁性海藻酸盐复合珠,应用于孔雀石绿降解,在酸性环境下磁性海藻酸盐复合珠对真菌有较好的保护作用,可应用于酸性条件下含染料废水的处理。Barapatre等[48]将黄曲霉菌(Aspergillus flavus)固定于5种惰性支撑材料上,固定化后的真菌对孔雀石绿降解率高达97%~99%,显著高于游离菌体。Alam等[94]利用黑曲霉菌(Aspergillus niger)固定于活性炭上,制备了一种去除染料的生物吸附剂。
Vršanská等[80]利用从云芝栓孔菌(Trametes versicolor)和木蹄层孔菌(Fomes fomentarius)体内提纯漆酶制备交联酶聚合体,与游离漆酶相比,在最佳条件下交联酶聚合体拥有更高降解率、稳定性和重复使用性,其中稳定性体现在pH值和温度稳定范围更广、储存稳定性更高、抗化学诱变能力更强。Yang等[83]采用了4种方式固定黑孔菌HYB07(Cerrena sp. HYB07)提纯漆酶,分别是海藻酸盐包封、壳聚糖共价结合、交联酶聚合体、磁性交联酶聚合体(M-CLEAs),表明固定化提高了反应温度和pH值稳定性。Wen等[84]将云芝栓孔菌(Trametes versicolor)提纯漆酶固定于高岭土上,在镉离子和3,5-二甲氧基-4-羟基苯甲醛(SA)共存条件下,有利于复合体对孔雀石绿脱色,或可应用于镉离子与孔雀石绿共存废水处理。Li等[87]将灵芝(Ganoderma lucidum)漆酶固定于菌丝体上,形成超细纤维组成白色膜状结构,具有良好的生物相容性,可用作药物载体。Huang等[93]将漆酶和介体固定于双层氢氧化物/藻酸盐生物杂化颗粒上,制备成了一种新型生物催化剂(Im-LMS),具有较强的可回收性和稳定性。
2.6 组学研究有关真菌降解孔雀石绿的组学研究较少,仅有Gomaa等[82]利用转录组学研究了氯化钙对黑曲霉菌(Aspergillus niger)降解孔雀石绿的分子调控机制。结果表明,在加入氯化钙30 min后,有28个相关基因被调控,从而增强了真菌对孔雀石绿的降解能力;此外,孔雀石绿降解过程与线粒体细胞色素C有关,细胞色素C氧化酶活性是孔雀石绿降解的关键因素。
3 结论与展望具有孔雀石绿降解能力的细菌共有30种,超过半数属于变形菌门;而真菌共有55种,超过半数属于担子菌门。真菌提纯酶降解孔雀石绿的最适温度较真菌菌体、细菌提纯酶与细菌菌体高,而最适pH值较它们都更小;即真菌提纯酶较真菌菌体、细菌提纯酶与细菌菌体更能适应高温偏酸性的降解环境。细菌与真菌对孔雀石绿的降解途径大致相同,均包括两个主要途径:N-去甲基化途径与中心碳原子羟基化途径;但细菌降解终产物多为线性化合物,而真菌降解终产物多包含苯环结构。细菌与真菌参与降解孔雀石绿的酶系均属漆酶系与锰过氧化物酶系;固定化细菌菌体、真菌菌体和相应提纯酶,均可提高降解率与稳定性。
截至目前,有关微生物降解孔雀石绿的组学研究依然十分匮乏。因此,利用多组学分析,可从基因组、转录组、蛋白质组、代谢组等多层次更深入地探索与阐释孔雀石绿生物降解的分子机制。可为孔雀石绿微生物降解机理研究、菌剂开发提供方法学支撑。
| [1] |
DONYA S M, FARGHALY A A, ABO-ZEID M A, et al. Malachite Green induces genotoxic effect and biochemical disturbances in mice[J]. European Review for Medical and Pharmacological Sciences, 2012, 16(4): 469-482. |
| [2] |
MITTELSTAEDT R A, MEI N, WEBB P J, et al. Genotoxicity of malachite green and leucomalachite green in female Big Blue B6C3F(1) mice[J]. Mutation Research-Genetic Toxicology and Environmental Mutagenesis, 2004, 561(1-2): 127-138. DOI:10.1016/j.mrgentox.2004.04.003 |
| [3] |
CULP S J, MELLICK P W, TROTTER R W, et al. Carcinogenicity of malachite green chloride and leucomalachite green in B6C3F(1) mice and F344 rats[J]. Food and Chemical Toxicology, 2006, 44(8): 1204-1212. DOI:10.1016/j.fct.2006.01.016 |
| [4] |
GUPTA S, SUNDARRAJAN M, RAO K V K. Tumor promotion by metanil yellow and malachite green during rat hepatocar-cinogenesis is associated with dysregulated expression of cell cycle regulatory proteins[J]. Teratogenesis Carcinogenesis and Mutagenesis, 2003, 301-312. |
| [5] |
崔松林. 辽宁省淡水鱼中孔雀石绿残留量检测[J]. 安徽农业科学, 2014, 42(13): 3916-3917. DOI:10.3969/j.issn.0517-6611.2014.13.055 |
| [6] |
华永有, 邱文倩, 周亮, 等. 市售淡水鱼中孔雀石绿及其代谢物残留量的调查研究[J]. 中国食品卫生杂志, 2011, 23(6): 563-566. |
| [7] |
刘书贵, 尹怡, 单奇, 等. 广东省鳜鱼和杂交鳢中孔雀石绿和硝基呋喃残留调查及暴露评估[J]. 中国食品卫生杂志, 2015, 27(5): 553-558. |
| [8] |
邵生文, 闻胜, 王艳, 等. 湖北省淡水鱼中孔雀石绿、结晶紫监测结果分析[J]. 公共卫生与预防医学, 2011, 22(2): 49-50. |
| [9] |
王敏娟, 聂晓玲, 程国霞, 等. 陕西省淡水鱼中孔雀石绿的污染调查及居民膳食暴露评估[J]. 卫生研究, 2015, 44(6): 965-969. |
| [10] |
张艺蓓, 岳田利, 乔海鸥, 等. 2013年陕西市售水产品中孔雀石绿和结晶紫的检测[J]. 西北农林科技大学学报(自然科学版), 2015, 43(5): 192-200. |
| [11] |
KWAN P P, BANERJEE S, SHARIFF M, et al. Quantitative analysis of malachite green and leucomalachite green residues in fish purchased from the markets in Malaysia[J]. Food Control, 2018, 92: 101-106. DOI:10.1016/j.foodcont.2018.04.031 |
| [12] |
陈文华, 李刚, 许方程, 等. 染料废水污染现状及处理方法研究进展[J]. 浙江农业科学, 2014(2): 264-269. DOI:10.3969/j.issn.0528-9017.2014.02.041 |
| [13] |
SANI R K, BANERJEE U C. Decolorization of triphenylmethane dyes and textile and dye-stuff effluent by Kurthia sp[J]. Enzyme and Microbial Technology, 1999, 24(7): 433-437. DOI:10.1016/S0141-0229(98)00159-8 |
| [14] |
DENG D, GUO J, ZENG G, et al. Decolorization of anthraquinone, triphenylmethane and azo dyes by a new isolated Ba-cillus cereus strain DC11[J]. International Biodeterioration & Biodegradation, 2008, 62(3): 263-269. |
| [15] |
GOMARE S S, PARSHETTI G K, GOVINDWAR S P. Biodegradation of malachite green by Brevibacillus laterosporus MTCC 2298[J]. Water Environment Research, 2009, 81(11): 2329-2336. DOI:10.2175/106143009X407357 |
| [16] |
AYED L, CHAIEB K, CHEREF A, et al. Biodegradation of triphenylmethane dye malachite green by Sphingomonas paucimobilis[J]. World Journal of Microbiology & Biotechnology, 2009, 25(4): 705-711. |
| [17] |
CHEN C Y, KUO J T, CHENG C Y, et al. Biological decolorization of dye solution containing malachite green by Pandoraea pulmonicola YC32 using a batch and continuous system[J]. Journal of Hazardous Materials, 2009, 172(2-3): 1439-1445. DOI:10.1016/j.jhazmat.2009.08.009 |
| [18] |
WU J, JUNG B G, KIM K S, et al. Isolation and charact-erization of Pseudomonas otitidis WL-13 and its capacity to decolorize triphenylmethane dyes[J]. Journal of Envir-onmental Sciences, 2009, 21(7): 960-964. DOI:10.1016/S1001-0742(08)62368-2 |
| [19] |
LI L T, HONG Q, YAN X, et al. Isolation of a malachite green-degrading Pseudomonas sp. MDB-1 strain and cloning of the tmr2 gene[J]. Biodegradation, 2009, 20(6): 768-776. |
| [20] |
CHEN C H, CHANG C F, LIU S M. Partial degradation mechanisms of malachite green and methyl violet B by Shewanella decolorationis NTOU1 under anaerobic conditions[J]. Journal of Hazardous Materials, 2010, 177(1-3): 281-289. DOI:10.1016/j.jhazmat.2009.12.030 |
| [21] |
DU L N, WANG S, LI G, et al. Biodegradation of malachite green by Pseudomonas sp. strain DY1 under aerobic condition: characteristics, degradation products, enzyme analysis and phytotoxicity[J]. Ecotoxicology, 2011, 20(2): 438-446. DOI:10.1007/s10646-011-0595-3 |
| [22] |
LV G Y, CHENG J H, CHEN X Y, et al. Biological decolorization of malachite green by Deinococcus radiodurans R1[J]. Bioresource Technology, 2013, 144: 275-280. DOI:10.1016/j.biortech.2013.07.003 |
| [23] |
SINIRLIOGLU Z A, SINIRLIOGLU D, AKBAS F. Preparation and characterization of stable cross-linked enzyme aggregates of novel laccase enzyme from Shewanella putr-efaciens and using malachite green decolorization[J]. Bioresource Technology, 2013, 146: 807-811. DOI:10.1016/j.biortech.2013.08.032 |
| [24] |
VIJAYALAKSHMIDEVI S R, MUTHUKUMAR K. Biodegradation of malachite green by Ochrobactrum sp[J]. World Journal of Microbiology & Biotechnology, 2014, 30(2): 429-437. |
| [25] |
MUKHERJEE T, DAS M. Degradation of malachite green by Enterobacter asburiae strain XJUHX-4TM[J]. Clean-Soil Air Water, 2014, 42(6): 849-856. DOI:10.1002/clen.201200246 |
| [26] |
MNIF I, FENDRI R, GHRIBI D. Malachite green bioremoval by a newly isolated strain Citrobacter sedlakii RI11; enhancement of the treatment by biosurfactant addition[J]. Water Science and Technology, 2015, 72(8): 1283-1293. DOI:10.2166/wst.2015.302 |
| [27] |
WANG R, REN C, HUANG N, et al. Draft genome sequence of Pseudomonas oleovorans strain MGY01 isolated from deep sea water[J]. Marine Genomics, 2015, 20: 17-18. DOI:10.1016/j.margen.2014.12.003 |
| [28] |
TAO Y, WANG F, MENG L, et al. Biological decolorization and degradation of malachite green by Pseudomonas sp. YB2: Process optimization and biodegradation pathway[J]. Current Microbiology, 2017, 74(10): 1210-1215. DOI:10.1007/s00284-017-1306-y |
| [29] |
GAN L, ZHOU F, OWENS G, et al. Burkholderia cepacia immobilized on eucalyptus leaves used to simultaneously remove malachite green(MG) and Cr(Ⅵ)[J]. Colloids and Surfaces B-Biointerfaces, 2018, 172: 526-531. DOI:10.1016/j.colsurfb.2018.09.008 |
| [30] |
LI B, GAN L, OWENS G, et al. New nano-biomaterials for the removal of malachite green from aqueous solution via a response surface methodology[J]. Water Research, 2018, 146: 55-66. DOI:10.1016/j.watres.2018.09.006 |
| [31] |
SHANG N, DING M, DAI M, et al. Biodegradation of malachite green by an endophytic bacterium Klebsiella aerogenes S27 involving a novel oxidoreductase[J]. Applied Microbiology and Biotechnology, 2019, 103(5): 2141-2153. DOI:10.1007/s00253-018-09583-0 |
| [32] |
MA X, LIU L, LI Q, et al. High-level expression of a bacterial laccase, CueO from Escherichia coli K12 in Pichia pastoris GS115 and its application on the decolorization of synthetic dyes[J]. Enzyme and Microbial Technology, 2017, 103: 34-41. DOI:10.1016/j.enzmictec.2017.04.004 |
| [33] |
AYED L, BAKIR K, BEN MANSOUR H, et al. In vitro mutagenicity, NMR metabolite characterization of azo and triphenylmethanes dyes by adherents bacteria and the role of the "cna" adhesion gene in activated sludge[J]. Microbial Pathogenesis, 2017, 103: 29-39. DOI:10.1016/j.micpath.2016.12.016 |
| [34] |
WANYONYI W C, ONYARI J M, SHIUNDU P M, et al. Biodegradation and detoxification of malachite green dye using novel enzymes from Bacillus cereus strain KM201428: Kinetic and metabolite analysis; proceedings of the international conference on technologies and materials for renewable energy, environment and sustainability(TMREES), Beirut, LEBANON, F 2017 Apr 21-24, 2017[C]. 2017.
|
| [35] |
甘莉, 路则洋, 林加奖, 等. EDTA增强Cr(Ⅵ)共存时Burkholderia cepacia降解孔雀绿的效率[J]. 环境科学学报, 2017, 37(12): 4610-4616. |
| [36] |
KOBAYASHI T, TAYA H, WILAIPUN P, et al. Malachite-green-removing properties of a bacterial strain isolated from fish ponds in Thailand[J]. Fisheries Science, 2017, 83(5): 827-835. DOI:10.1007/s12562-017-1102-4 |
| [37] |
李刚, 都林娜, 许方程, 等. 肠杆菌CV-b脱色孔雀石绿的特性及机制[J]. 浙江大学学报(农业与生命科学版), 2017, 43(4): 493-501. |
| [38] |
FARHA A K, THASNEEM T R, PURUSHOTHAMAN A, et al. Phylogenetic diversity and biotechnological potentials of marine bacteria from continental slope of eastern Arabian Sea[J]. Journal of Genetic Engineering and Biotechnology, 2018, 16(2): 253-258. DOI:10.1016/j.jgeb.2018.06.002 |
| [39] |
QU W, HONG G, ZHAO J. Degradation of malachite green dye by Tenacibaculum sp. HMG1 isolated from Pacific deepsea sediments[J]. Acta Oceanologica Sinica, 2018, 37(6): 104-111. DOI:10.1007/s13131-018-1187-3 |
| [40] |
刘单单, 李春生, 杨贤庆, 等. 孔雀石绿降解菌Enterobacter sp. B-20的分离、鉴定和降解特性研究[J]. 南方水产科学, 2018, 14(1): 50-59. |
| [41] |
QIAO W, LIU H. Enhanced decolorization of malachite green by a magnetic graphene oxide-CotA laccase composite[J]. International Journal of Biological Macromolecules, 2019, 138: 1-12. DOI:10.1016/j.ijbiomac.2019.07.077 |
| [42] |
LI C, LIU D, YANG X, et al. Efficient biodegradation of malachite green by a newly isolated Klebsiella pneumo-niae strain WA-1[J]. Environmental Progress & Sustainable Energy, 2020, 39(4): e13346. |
| [43] |
SUTAR S S, PATIL P J, TAMBOLI A S, et al. Biodegradation and detoxification of malachite green by a newly isolated bioluminescent bacterium Photobacterium leiognathi strain MS under RSM optimized culture conditions[J]. Biocatalysis and Agricultural Biotechnology, 2019, 20: 101183. DOI:10.1016/j.bcab.2019.101183 |
| [44] |
VIGNESH A, MANIGUNDAN K, SANTHOSHKUMAR J, et al. Microbial degradation, spectral analysis and toxicological assessment of malachite green by Streptom-yces chrest-omyceticus S20[J]. Bioprocess and Biosystems Engine-ering, 2020, 43(8): 1457-1468. DOI:10.1007/s00449-020-02339-z |
| [45] |
ASADI E, MAKHDOUMI A, ASOODEH A. Laccase mediator system obtained from a marine spore exhibits decolorization potential in harsh environmental conditions[J]. Ecotoxicology and Environmental Safety, 2020, 191: 110184. DOI:10.1016/j.ecoenv.2020.110184 |
| [46] |
SONG J, HAN G, WANG Y, et al. Pathway and kinetics of malachite green biodegradation by Pseudomonas veronii[J]. Scientific Reports, 2020, 10(1): 4502. DOI:10.1038/s41598-020-61442-z |
| [47] |
QU W, LIU T, WANG D, et al. Metagenomics-based discovery of malachite green-degradation gene families and enzymes from mangrove sediment[J]. Frontiers in Microbiology, 2018(9): 2187. |
| [48] |
BARAPATRE A, AADIL K R, JHA H. Biodegradation of malachite green by the ligninolytic fungus Aspergillus flavus[J]. Clean-Soil Air Water, 2017, 45(4): 1600045. DOI:10.1002/clen.201600045 |
| [49] |
BUMPUS J A, BROCK B J. Biodegradation of crystal violet by the white rot fungus Phanerochaete chrysosporium[J]. Applied and Environmental Microbiology, 1988, 54(5): 1143-1150. DOI:10.1128/aem.54.5.1143-1150.1988 |
| [50] |
LEVIN L, PAPINUTTI L, FORCHIASSIN F. Evaluation of Argentinean white rot fungi for their ability to produce ligninmodifying enzymes and decolorize industrial dyes[J]. Bioresource Technology, 2004, 94(2): 169-176. DOI:10.1016/j.biortech.2003.12.002 |
| [51] |
PAPINUTTI V L, FORCHIASSIN F. Modification of mala-chite green by Fomes sclerodermeus and reduction of toxicity to Phanerochaete chrysosporium[J]. Fems Microbiology Letters, 2004, 231(2): 205-209. DOI:10.1016/S0378-1097(03)00957-1 |
| [52] |
EICHLEROVA I, HOMOLKA L, NERUD F. Synthetic dye decolorization capacity of white rot fungus Dichomitus squalens[J]. Bioresource Technology, 2006, 97(16): 2153-2159. DOI:10.1016/j.biortech.2005.09.014 |
| [53] |
EICHLEROVA I, HOMOLKA L, NERUD F. Evaluation of synthetic dye decolorization capacity in Ischnoderma resinosum[J]. Journal of Industrial Microbiology & Biotechnology, 2006, 33(9): 759-766. |
| [54] |
PAPINUTTI L, MOUSO N, FORCHIASSIN F. Removal and degradation of the fungicide dye malachite green from aqueous solution using the system wheat bran-Fomes scleroder-meus[J]. Enzyme and Microbial Technology, 2006, 39(4): 848-853. DOI:10.1016/j.enzmictec.2006.01.013 |
| [55] |
PANT D, SINGH A, SATYAWALI Y, et al. Effect of carbon and nitrogen source amendment on synthetic dyes decolourizing efficiency of white-rot fungus, Phanerochaete chrysosporium[J]. Journal of Environmental Biology, 2008, 29(1): 79-84. |
| [56] |
JAYASINGHE C, IMTIAJ A, LEE G W, et al. Degradation of three aromatic dyes by white rot fungi and the production of ligninolytic enzymes[J]. Mycobiology, 2008, 36(2): 114-120. DOI:10.4489/MYCO.2008.36.2.114 |
| [57] |
MURUGESAN K, YANG I-H, KIM Y-M, et al. Enhanced transformation of malachite green by laccase of Ganoderma luc-idum in the presence of natural phenolic compounds[J]. Applied Microbiology and Biotechnology, 2009, 82(2): 341-350. DOI:10.1007/s00253-008-1819-1 |
| [58] |
MAALEJ-KAMMOUN M, ZOUARI-MECHICHI H, BELBAHRI L, et al. Malachite green decolourization and detoxification by the laccase from a newly isolated strain of Trametes sp[J]. International Biodeterioration & Biodegradation, 2009, 63(5): 600-606. |
| [59] |
CHHABRA M, MISHRA S, SREEKRISHNAN T R. Laccase/mediator assisted degradation of triarylmethane dyes in a continuous membrane reactor[J]. Journal of Biotec-hnology, 2009, 143(1): 69-78. DOI:10.1016/j.jbiotec.2009.06.011 |
| [60] |
LEVIN L, MELIGNANI E, MARCELA RAMOS A. Effect of nitrogen sources and vitamins on ligninolytic enzyme production by some white-rot fungi. Dye decolorization by selected culture filtrates[J]. Bioresource Technology, 2010, 101(12): 4554-4563. DOI:10.1016/j.biortech.2010.01.102 |
| [61] |
FOROOTANFAR H, FARAMARZI M A, SHAHVERDI A R, et al. Purification and biochemical characterization of extracellular laccase from the ascomycete Paraconiothyrium variabile[J]. Bioresource Technology, 2011, 102(2): 1808-1814. DOI:10.1016/j.biortech.2010.09.043 |
| [62] |
GRASSI E, SCODELLER P, FILIEL N, et al. Potential of Trametes trogii culture fluids and its purified laccase for the decolorization of different types of recalcitrant dyes without the addition of redox mediators[J]. International Biod-eterioration & Biodegradation, 2011, 65(4): 635-643. |
| [63] |
BIBI I, BHATTI H N, ASGHER M. Comparative study of natural and synthetic phenolic compounds as efficient laccase mediators for the transformation of cationic dye[J]. Biochemical Engineering Journal, 2011, 56(3): 225-231. DOI:10.1016/j.bej.2011.07.002 |
| [64] |
LEVIN L, DIORIO L, GRASSI E, et al. Grape stalks as substrate for white rot fungi, lignocellulolytic enzyme production and dye decolorization[J]. Revista Argentina De Microbiologia, 2012, 44(2): 105-112. |
| [65] |
BALAN K, SATHISHKUMAR P, PALVANNAN T. Decolorization of malachite green by laccase: Optimization by response surface methodology[J]. Journal of the Taiwan Institute of Chemical Engineers, 2012, 43(5): 776-782. DOI:10.1016/j.jtice.2012.04.005 |
| [66] |
SARAVANAKUMAR K, KATHIRESAN K. Bioremoval of the synthetic dye malachite green by marine Trichoderma sp[J]. Springerplus, 2014(3): 631. |
| [67] |
RANI B, KUMAR V, SINGH J, et al. Bioremediation of dyes by fungi isolated from contaminated dye effluent sites for bio-usability[J]. Brazilian Journal of Microbiology, 2014, 45(3): 1055-1063. |
| [68] |
YANG C H, SHIH M C, CHIU H C, et al. Magnetic Pycnoporus sanguineus-loaded alginate composite beads for removing dye from aqueous solutions[J]. Molecules, 2014, 19(6): 8276-8288. |
| [69] |
YAN J, NIU J, CHEN D, et al. Screening of Trametes strains for efficient decolorization of malachite green at high temperatures and ionic concentrations[J]. International Biodeterioration & Biodegradation, 2014, 87: 109-115. |
| [70] |
JASINSKA A, PARASZKIEWICZ K, SIP A, et al. Malachite green decolorization by the filamentous fungus Myrothecium roridum-Mechanistic study and process optimiza-tion[J]. Bioresource Technology, 2015, 194: 43-48. |
| [71] |
CHEN S H, SU A, TING Y. Biodecolorization and biodegradation potential of recalcitrant triphenylmethane dyes by Coriolopsis sp. isolated from compost[J]. Journal of Environmental Management, 2015, 150: 274-280. |
| [72] |
YANG J, YANG X, LIN Y, et al. Laccase-catalyzed decolorization of malachite green: performance Optimization anddegradation mechanism[J]. Plos One, 2015, 10(5): e0-127714. |
| [73] |
LING Z R, WANG S S, ZHU M J, et al. An extracellular laccase with potent dye decolorizing ability from white rot fungus Trametes sp. LAC-01[J]. International Journal of Biological Macromolecules, 2015, 81: 785-793. |
| [74] |
SHARMA A, SHRIVASTAVA B, KUHAD R C. Reduced toxicity of malachite green decolorized by laccase produced from Ganoderma sp. rckk-02 under solid-state fermentation[J]. 3 Biotech, 2015, 5(5): 621-631. |
| [75] |
YANG H, SUN H, ZHANG S, et al. Potential of acetylacetone as a mediator for Trametes versicolor laccase in enzymatic transformation of organic pollutants[J]. Environmental Sci-ence and Pollution Research, 2015, 22(14): 10882-10889. |
| [76] |
CHMELOVA D, ONDREJOVIC M. Purification and characterization of extracellular laccase produced by Ceriporiopsis subvermispora and decolorization of triphenylmethane dyes[J]. Journal of Basic Microbiology, 2016, 56(11): 1173-1182. |
| [77] |
WANG B, YAN Y, TIAN Y, et al. Heterologous expression and characterisation of a laccase from Colletotrichum lagenarium and decolourisation of different synthetic dyes[J]. World Journal of Microbiology & Biotechnology, 2016, 32(3): 40. |
| [78] |
YANG X, ZHENG J, LU Y, et al. Degradation and detoxification of the triphenylmethane dye malachite green catalyzed by crude manganese peroxidase from Irpex lacteus F17[J]. Environmental Science and Pollution Research, 2016, 23(10): 9585-9597. |
| [79] |
YUAN X, TIAN G, ZHAO Y, et al. Degradation of dyes using crude extract and a thermostable and pH-stable laccase isolated from Pleurotus nebrodensis[J]. Bioscience Reports, 2016, 36: e00365. |
| [80] |
VRANSKÁ M, VOBERKOVA S, JIMENEZ JIMENEZ A M, et al. Preparation and optimisation of cross-linked enzyme aggregates using native isolate white rot fungi Trametes versicolor and Fomes fomentarius for the decolourisation of synthetic dyes[J]. International Journal of Environmental Research and Public Health, 2018, 15(1): 23. |
| [81] |
MORALES-ALVAREZ E D, RIVERA-HOYOS C M, POVEDA-CUEVAS S A, et al. Malachite green and crystal violet dec-olorization by Ganoderma lucidum and Pleurotus ostreatus supe-rnatant and by rGlLCC1 and rPOXA 1B concentrates: Molecular docking analysis[J]. Applied Biochemistry and Biotechnology, 2018, 184(3): 794-805. |
| [82] |
GOMAA O M, SELIM N S, WEE J, et al. RNA Seq analysis of the role of calcium chloride stress and electron transport in mitochondria for malachite green decolorization by Aspergillus niger[J]. Fungal Genetics and Biology, 2017, 105: 1-7. |
| [83] |
YANG J, WANG Z, LIN Y, et al. Immobilized Cerrena sp. laccase: preparation, thermal inactivation, and operational stability in malachite green decolorization[J]. Scientific Reports, 2017(7): 712-730. |
| [84] |
WEN X, DU C, WAN J, et al. Immobilizing laccase on kaolinite and its application in treatment of malachite green effluent with the coexistence of Cd(Ⅱ)[J]. Chemosphere, 2019, 217: 843-850. |
| [85] |
WANG S N, CHEN Q J, ZHU M J, et al. An extracellular yellow laccase from white rot fungus Trametes sp. F1635 and its mediator systems for dye decolorization[J]. Biochimie, 2018, 148: 46-54. |
| [86] |
QIAO W, ZHANG Y, XIE Z, et al. Toxicity of perfluorooctane sulfonate on Phanerochaete chrysosporium: Growth, pollutant degradation and transcriptomics[J]. Ecotoxicology and Environmental Safety, 2019, 174: 66-74. |
| [87] |
LI G, WANG Q, LV P, et al. Bioremediation of dyes using ultrafine membrane prepared from the waste culture of Ganoderma lucidum with in-situ immobilization of laccase[J]. Bioresources, 2016, 11(4): 9162-9174. |
| [88] |
MORALES-ALVAREZ E D, RIVERA-HOYOS C M, CHAP-ARRO-NUNEZ L E, et al. Decolorization and deto-xification of malachite green by Ganoderma lucidum: Key operating parameters and adsorption studies[J]. Journal of Environmental Engineering, 2017, 143(4): 04016093. |
| [89] |
SHANMUGAM S, ULAGANATHAN P, SWAMINATHAN K, et al. Enhanced biodegradation and detoxification of malachite green by Trichoderma asperellum laccase: Degradation pathway and product analysis[J]. International Biodeterioration & Biodegradation, 2017, 125: 258-268. |
| [90] |
HUSSAIN S, QUINN L, LI J, et al. Simultaneous removal of malachite green and hexavalent chromium by Cunninghamella elegans biofilm in a semi-continuous system[J]. International Biodeterioration & Biodegradation, 2017, 125: 142-149. |
| [91] |
MARCHARCHAND S, TING A S Y. Trichoderma asperellum cultured in reduced concentrations of synthetic medium retained dye decolourization efficacy[J]. Journal of Env-ironmental Management, 2017, 203: 542-549. |
| [92] |
郑红叶, 薛雅蓉, 刘常宏. 裂褶菌cfcc7252菌株对孔雀石绿染料的高效降解[J]. 微生物学通报, 2017, 44(1): 38-48. |
| [93] |
HUANG J, YANG Y, WANG Y, et al. Immobilization of a laccase/2, 2'-azino-bis-(3-ethylbenzothiazoline)-6-sulfonic acid system to layered double hydroxide/alginate biohybrid beads for biodegradation of malachite green dye[J]. Biomed Research International, 2018, 2018(1): 471961. |
| [94] |
ALAM M Z, KHAN M J H, KABBASHI N A, et al. Development of an effective biosorbent by fungal immobilization technique for removal of dyes[J]. Waste and Biomass Valorization, 2018, 9(4): 681-690. |
| [95] |
CARDOSO B K, LINDE G A, COLAUTO N B, et al. Panus strigellus laccase decolorizes anthraquinone, azo, and triphenylmethane dyes[J]. Biocatalysis and Agricultural Bio-technology, 2018, 16: 558-563. |
| [96] |
RANIMOL G, VENUGOPAL T, GOPALAKRISHNAN S, et al. Production of laccase from Trichoderma harzianum and its application in dye decolourisation[J]. Biocatalysis and Agricultural Biotechnology, 2018, 16: 400-404. |
| [97] |
PANDEY R K, TEWARI S, TEWARI L. Lignolytic mushroom Lenzites elegans WDP2: Laccase production, characterization, and bioremediation of synthetic dyes[J]. Eco-toxicology and Environmental Safety, 2018, 158: 50-58. |
| [98] |
LALLAWMSANGA, LEO V V, PASSARI A K, et al. Elevated levels of laccase synthesis by Pleurotus pulmonarius BPSM10 and its potential as a dye decolorizing agent[J]. Saudi Journal of Biological Sciences, 2019, 26(3): 464-468. |
| [99] |
ZHUO R, ZHANG J, YU H, et al. The roles of Pleurotus ostreatus HAUCC 162 laccase isoenzymes in decolorization of synthetic dyes and the transformation pathways[J]. Chemosphere, 2019, 234: 733-745. |
| [100] |
ARUNPRASATH T, SUDALAI S, MEENATCHI R, et al. Biodegradation of triphenylmethane dye malachite green by a newly iso lated fungus strain[J]. Biocatalysis and Agricultural Biotechnology, 2019, 17: 672-679. |
| [101] |
CHEN S H, CHEOW Y L, NG S L, et al. Biodegradation of triphenylmethane dyes by non-white rot fungus Penicillium simplicissimum: Enzymatic and toxicity studies[J]. International Journal of Environmental Research, 2019, 13(2): 273-282. |
| [102] |
NEJAD Z G, BORGHEI S M, YAGHMAEI S. Biodegradation of synthetic dye using partially purified and characterized laccase and its proposed mechanism[J]. International Journal of Environmental Science and Technology, 2019, 16(12): 7805-7816. |
 2024, Vol. 16
2024, Vol. 16 

