内分泌干扰效应化合物(EDCs)是一类可能干扰生物体正常激素功能的化学物质[1]。根据特征官能团及其应用领域,EDCs一般可分为含酚羟基/羰基的激素和类固醇、农药和除草剂(含有氯取代基的芳香环和杂环)、个人护理用品添加剂(对羟基苯甲酸酯和苯酚)、增塑剂(双酚和邻苯二甲酸酯)和其他有机污染物等[2]。环境介质中的EDCs主要来源于人体和牲畜的排泄[3]。经管网收集,EDCs会汇入污水处理厂。作为水环境中EDCs的主要来源,污水中EDCs的质量浓度一般在ng/L到μg/L量级[4]。研究表明,环境中的EDCs可造成一系列生态风险并危害人体健康,例如导致生殖障碍、发育异常及引发癌症[5],破坏野生动物及人体内分泌系统正常功能[6]。因此,加强污水中EDCs的管控至关重要。
污水中EDCs种类繁多、结构复杂,对EDCs类化合物结构的识别和认知是对其进行风险评估和管控的前提。对于人类来说,EDCs主要存在于水源、土壤和沉积物中。某些EDCs高度溶于水,例如氟西汀和左炔诺孕酮等物质不受废水处理过程的影响,可以完好无损地释放到环境中。某些EDCs(苯并[a]芘和三氯生等)易与土壤和沉积物结合,从而在生物体中聚集,但土壤和沉积物中的EDCs浓度往往远低于水中的EDCs浓度[7]。环境中EDCs测定的前处理方法主要为:固相萃取(SPE)、固相微萃取(SPME)和液相微萃取(LPME)等[8]。环境介质中的EDCs浓度一般较低,液相色谱-串联高分辨质谱(LC-HRMS)是对其进行测定的最常用方法[9]。由于环境样品基质复杂,经LC-HRMS检测后,每个样品一般含有上万个特征峰[10]。因此,需要从这上万个特征峰中识别出具有内分泌干扰效应的特征峰或者化合物,从而鉴定环境样品中的EDCs。自上而下和自下而上策略是识别环境复杂介质中EDCs质谱信号并进行结构鉴定的2种常用策略[11-13]。自上而下策略是指,全面识别出这上万个特征峰所代表的化合物结构,进而根据毒性测试或计算毒理学方法获得所有化合物的内分泌干扰特性,最终识别出EDCs;自下而上策略的方法流程是,先通过分级分馏或机器学习模型预测的方式,识别出具有内分泌干扰效应的馏分或特征峰,进而对其代表的化合物进行结构鉴定,识别出环境样品中的EDCs[12]。
1 基于自上而下策略的环境样品中EDCs的识别方法研究进展自上而下策略的环境样品中EDCs识别方法流程主要包括:化合物结构识别和内分泌干扰效应测试。其中,基于高分辨质谱的化合物结构识别是主要难点[14-15],因此下文主要针对化合物结构识别进行阐述。采用高分辨质谱进行环境样品中EDCs结构识别的流程包括:样品采集、样品前处理、样品检测、高分辨质谱数据解析[14]。前3个流程目前已形成标准化步骤,难度较小,基于高分辨质谱数据进行化合物结构鉴定为主要难点。一般来说,可疑物筛查和非靶向筛查是进行环境样品中未知结构化合物识别的主要方法。针对EDCs,通过特异性前处理手段对其进行富集更有利于后续的可疑物筛查和非靶向筛查过程中的结构识别。目前,北京大学胡剑英[16]团队开发了多款基于内分泌干扰效应受体的蛋白萃取柱,可以有针对性地富集EDCs,从而极大简化了后续的数据处理流程。
1.1 环境样品中EDCs可疑物筛查方法研究进展环境样品中EDCs可疑物筛查是指,通过编制EDCs可疑物清单,进而将清单中化合物的理论质荷比与高分辨质谱检测的样品数据进行比对,以识别出污水样品质谱数据中可能存在的EDCs特征峰,进而结合保留时间和二级谱图数据对该色谱峰所代表的化合物结构进行鉴定和确证[17-18]。因此,编制EDCs可疑物清单是可疑物筛查最重要的环节。
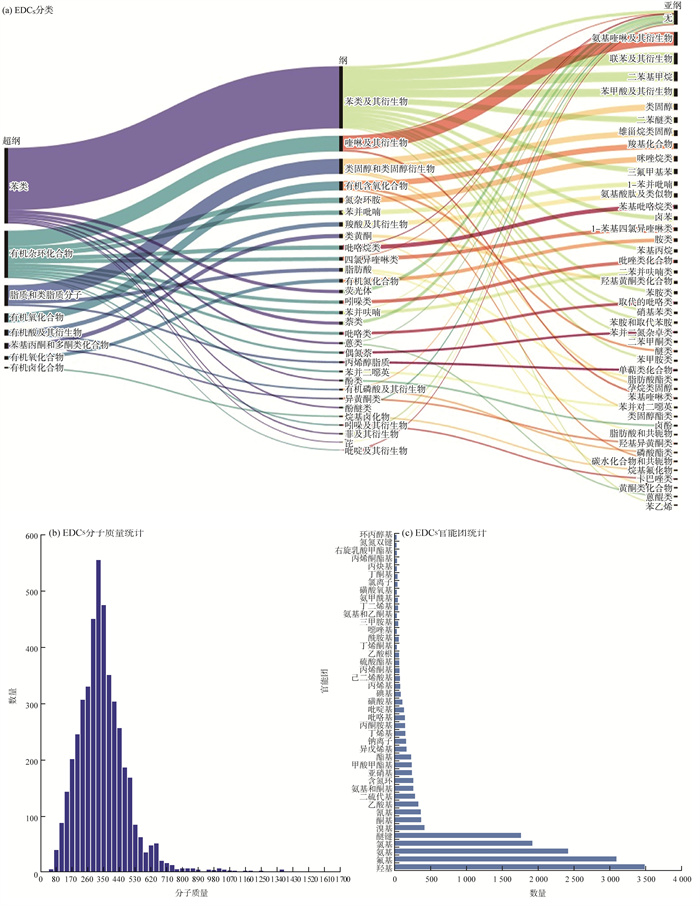
化合物毒性效应数据库和文献检索是获取EDCs最常用的方法[19]。通过对ToxCast数据库和文献中基于毒性测试和计算毒理学获取的EDCs进行检索[20-22],共获得包含4 512种EDCs的可疑物清单。结构分类结果表明,该清单中超纲、纲、亚纲层级化合物数量分别为8,30和44个[图 1(a)]。化合物的分子质量为30~1 701,其中,290~320,320~350范围内的化合物占比较大,分别为苯类和有机杂环化合物[图 1(b)],表明EDCs一般具有中等分子质量。
|
图 1 4 512种EDCs的特征统计 |
其中,苯类化合物构成了一个重要的研究分支,其主要成分包括苯及其衍生物。这些衍生物可进一步细分为联苯甲烷及其衍生物、二苯基甲烷、苯甲酸及其衍生物、二苯醚类化合物以及三氟甲基苯等。有机杂环化合物则是指含有杂原子的环状化合物,其中喹啉及其衍生物、偶氮烷烃和苯并吡喃等类型尤为突出。在这些化合物中,喹啉及其衍生物尤以氨基喹啉及其衍生物为代表。脂质和类脂质分子主要包括类固醇及其衍生物,其中雌甾烯类固醇和雄甾烷类固醇是类固醇衍生物的主要类型。在“超纲”水平上,苯类和有机杂环化合物占比最大,分别为25.9%和16.2%。在“纲”水平上,苯类及其衍生物占比最大,达21.4%。由上可见,应加强关注环境样品中具有苯环和杂环结构化合物的内分泌干扰效应。进一步对EDCs官能团特征进行分析[图 1(c)],发现羟基、氟基、氨基、氯基是EDCs最常见的官能团,占比分别为19.7%,17.5%,13.7%和10.9%。一般来说,化合物导致内分泌干扰效应的内在机制在于这些化合物具有某些结构特征,使其更容易与雌激素受体、雄激素受体或甲状腺激素受体结合,从而造成内分泌干扰效应[23]。因此,具有羟基、氟基、氨基和氯基官能团可能是化合物与不同受体结合的子结构,应尤其关注具有这些官能团结构特征的化合物。
通过可疑物筛查策略,研究者已识别出众多内分泌干扰效应化合物(表 1)。例如,Guo等[21]通过建立包含2 003个EDCs的可疑物清单,识别出地表水中39个EDCs,并发现己烯雌酚是地表水中雌激素效应的主要贡献者(贡献高达54%)。Muschket等[24]构建了包含302个EDCs的可疑物清单,并对污水样品中的EDCs进行识别。可能是由于所构建的清单包含的化合物数目较小,在污水样品中未识别出该清单中的化合物。这表明,所构建的EDCs清单数目是影响可疑物筛查所得EDCs数目的关键[18]。此外,可疑物筛查清单中的化合物均为已知化合物,因此,污水样品中结构未知的EDCs有待使用其他方法(如非靶向筛查方法)进行识别。
| 表 1 水环境中可疑物筛查方法识别EDCs案例 |
环境样品中EDCs非靶向筛查是指,事先不预设可能存在的EDCs,通过直接对高分辨质谱数据进行解析,识别出可能存在的EDCs[27-28]。由于环境样品基质复杂,单个样品所含的质谱信号一般高达上万个,这使得基于非靶向策略的EDCs识别极为困难。有效的质谱数据非靶向分析工具可以部分解决化合物识别的难题,简化数据解析流程[29-30]。
高分辨质谱数据的非靶向峰提取是开展非靶向筛查的第一步。目前开源的非靶向峰提取工具有MS-DIAL、MZmine2、XCMS、enviMass等[31-34]。经过背景扣除、非靶向峰提取、锋对齐等操作,环境样品一般可以提取上万甚至数万个色谱峰信号。值得注意的是,这些色谱峰信号与化合物物质信号并非一一对应。一般来说,化合物经质谱碎裂后会发生源内裂解、聚合,加不同离子(氢离子、钠离子、钾离子)生成不同的准分子离子峰。另外,由于同一元素存在不同的同位素,这在质谱中也会形成一个化合物对应多个不同同位素造成的色谱峰信号[35]。因此,高分辨质谱全扫描获得的质谱信号一般存在较大冗余,去冗余可以为后续的结构解析减轻工作量。MS-DIAL、MZmine2软件可以进行同位素、不同加和离子的确认,但对源内裂解、聚合物等形式的色谱峰难以甄别[32, 36]。其他一些软件或R、Python包可以有效对源内裂解、聚合峰进行去冗余。例如,大连化学物理研究所许国旺团队开发的离子融合方法可以有效识别源内裂解峰,从而将原始色谱峰从609和1 084个削减至106和169个[37]。R包CROP可以有效扣除源内裂解峰,减少98%的冗余峰信息[38]。
提取出的色谱峰需要进一步进行结构识别。由于环境样品中存在大量未知的化合物,其结构鉴定尤为困难,有效的质谱数据组织方式可以辅助未知化合物的结构鉴定。分子网络方法是近年非靶向筛查领域广泛使用的质谱数据组织方法[10]。其原理在于,结构类似的化合物一般具有类似的二级谱图。因此可以通过谱图的类似性将结构类似的化合物聚到网络图中的同一个簇或相邻节点,从而通过网络中已知化合物的结构辅助其相邻未知结构化合物的鉴定[39-40],目前使用该方法已识别出一系列转化产物、天然产物等未知化合物[41-43]。另外,研究发现,分子网络所聚类到同一个簇的结构类似化合物往往具有类似的生物活性[44],因此可以基于分子网络对质谱数据进行前处理,辅助环境复杂样品中EDCs质谱信号的识别,从而进行结构鉴定。
通过非靶向筛查方法,研究者已识别出一系列EDCs(表 2)。例如,Muschket等[24]采用MetFrag等非靶向筛查工具,识别出污水样品中3个未知的EDCs。Corine等[47]通过Metaboscape等非靶向筛查工具,识别出污水样品中7个EDCs,并发现杀虫剂异甲草胺和氰唑胺是地表水中抗孕激素和抗雄激素活性的主要来源。与可疑物筛查方法相比,非靶向筛查方法所识别出的EDCs数目明显更少,这是由于非靶向筛查方法数据处理更繁琐,数据分析难度更大。但该方法有利于识别出未知的EDCs。因此,在未来研究中,可将可疑物筛查和非靶向筛查方法结合,以便更有效地识别出环境中的EDCs。
| 表 2 水环境中非靶向筛查方法识别EDCs案例 |
自下而上策略的环境样品中EDCs识别方法主要包括:具有内分泌干扰效应的质谱信号识别和该质谱信号所代表的化合物结构识别与鉴定。由于化合物结构鉴定方法在自上而下策略的筛查策略中已介绍,这里重点阐述具有内分泌干扰效应的质谱信号精准识别方法。一般来说,基于效应导向分析(EDA)是最常用的识别质谱信号中具有内分泌干扰效应信号或组分的方法,且已得到广泛应用[27]。另外,近年随着机器学习算法及大数据等的发展,基于模型直接对质谱谱图进行预测以获得内分泌干扰效应化合物质谱信号的方法已得到应用[48-49]。
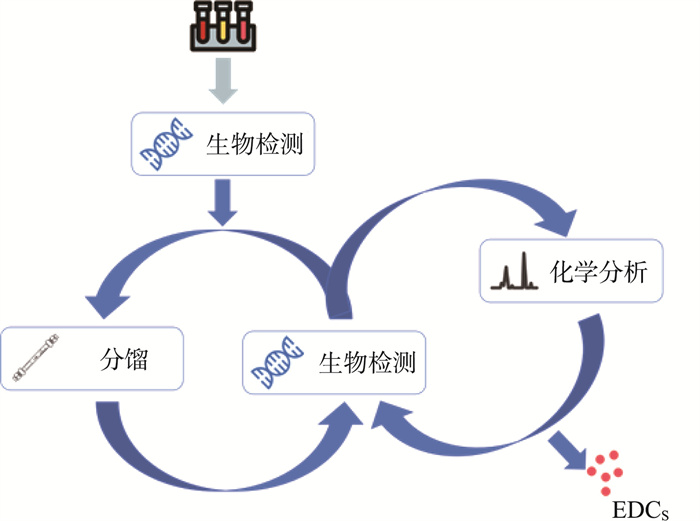
2.1 基于EDA策略的环境样品中EDCs识别基于EDA策略的环境样品中EDCs识别流程见图 2。由图 2可见,EDA方法结合了化学分析与生物检测方法,采用色谱柱分离不同极性化合物,实现不同辛醇水分配系数化合物的分组,获得不同组分的样品,进一步对不同组分样品进行测试,从而确认哪个组分具有内分泌干扰效应,进而对具有内分泌干扰效应组分中的质谱信号进行结构识别与鉴定,以识别出EDCs[28]。
|
图 2 基于EDA策略的环境样品中EDCs识别流程 |
目前研究者通过EDA方法已识别出一系列EDCs(表 3)。例如,Muschket等[24]采用不同色谱柱(C18、PFP、NH2、PYE)对污水样品进行分离,进而通过抗雄激素CALUX测试方法识别内分泌干扰效应活性组分,最后通过液相色谱-高分辨质谱-串联质谱非靶向筛查、质谱碎片离子预测等方式鉴定化合物,确定了4-甲基-7-二乙基氨基香豆素(C47)及其2种衍生物是污水样品中高抗雄激素类化合物。Zwart等[50]通过报告基因检测方法,对色谱分离的228个组分进行活性检测,并结合非靶向识别分析,发现8种化合物,并进一步确认了其中2种(1,2,3-苯并三氮唑和雄烯二酮)分别为活性致突变物和雄激素化合物。Zwart等[45]对污水厂水样进行报告基因检测,从而识别出13种可能的EDCs,进一步通过生物测试验证确证了其中10种化合物具有内分泌干扰效应,并发现二氧化苯和胡椒碱生物活性贡献度最高。Lopez-Herguedas等[52]采用自动现场大体积固相萃取法,收集医院废水样品,并采用正交色谱分离和A-YES生物分析法测定不同组分的生物活性,发现医院废水表现出显著的雌激素活性,在浓缩倍数为1时,其雌激素活性为11 ng EEQ/L(每升水中化学物质的毒性等效质量)。雌激素活性贡献最高的化合物是美司他诺、雌酮、双酚A和丁基对羟基苯甲酸酯,其雌激素活性贡献率>50%。
| 表 3 EDCs的EDA识别案例 |
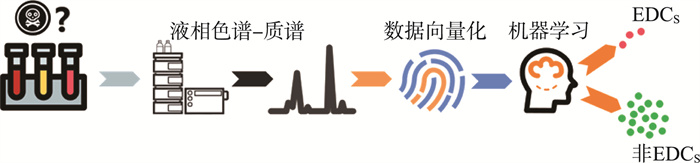
基于谱图机器学习模型分类的EDCs识别流程见图 3。由图 3可见,该流程直接对质谱信号(二级谱图)进行二分类机器学习模型构建,从而对污水样品质谱信号进行预测,识别出EDCs的质谱信号,进一步对其进行结构鉴定,识别出污水中的EDCs[48]。该方法的关键在于建立预测效果准确的质谱信号与内分泌干扰效应间关系的二分类模型。建立该模型过程中,获取EDCs的质谱二级谱图对模型预测效果起主要作用。目前,Tox21数据库中包含11 764种EDCs,MassBank质谱谱图数据库中包含上百万张化合物二级谱图[54]。以Tox21数据库中有明确内分泌干扰效应测试结果的化合物[55],进一步搜集这些化合物的二级谱图作为训练集,并结合不同机器学习算法,如随机森林、支持向量机、图卷积等进行模型构建,以准确预测未知质谱信号是否代表EDCs。目前已有研究者基于Tox21的EDCs和MassBank质谱二级谱图数据作为训练集,构建了基于随机森林、支持向量机、逻辑斯回归、K近邻等算法的谱图机器学习模型,其中最好的模型效果平均ROC-AUC(ROC曲线和AUC值,用于评估二分类模型的整体性能)得分为0.75。基于谱图机器学习模型,可以将识别EDCs的质谱信号削减75%,极大地减少了复杂样品中EDCs的识别工作[12]。
|
图 3 基于谱图机器学习模型分类的EDCs识别流程 |
基于目前EDCs识别方法的问题,提出以下展望:(1)进一步扩充EDCs结构清单和质谱二级谱图清单,以实现更高通量、快速的环境样品中EDCs可疑物与非靶向筛查。(2)完善EDCs质谱二级谱图到活性分类模型,并构建基于该模型的应用工具和平台,辅助研究人员快速识别环境样品中的EDCs。
| [1] |
杨廷政, 桑宇驰, 吕禾源, 等. 环境中典型内分泌干扰物(EDCs)去除技术研究进展[J]. 当代化工研究, 2024(3): 6-8. |
| [2] |
WANG R, MA X, LIU T, et al. Degradation aspects of endocrine disrupting chemicals: A review on photocatalytic processes and photocatalysts[J]. Applied Catalysis A: General, 2020, 597: 117547. DOI:10.1016/j.apcata.2020.117547 |
| [3] |
ZHONG R, ZOU H, GAO J, et al. A critical review on the distribution and ecological risk assessment of steroid hormones in the environment in China[J]. Science of The Total Environment, 2021, 786: 147452. DOI:10.1016/j.scitotenv.2021.147452 |
| [4] |
XIAO Y, HAN D, CURRELL M, et al. Review of endocrine disrupting compounds(EDCs) in China's water environments: Implications for environmental fate, transport and health risks[J]. Water Research, 2023, 245: 120645. DOI:10.1016/j.watres.2023.120645 |
| [5] |
邓茂先, 陈祥贵. 环境内分泌干扰物研究进展[J]. 国外医学(卫生学分册), 2000(2): 65-68, 77. |
| [6] |
鄂勇, 张晓琳, 宋秋霞. 环境内分泌干扰物及其潜在威胁[J]. 东北农业大学学报, 2008, 39(11): 135-139. DOI:10.3969/j.issn.1005-9369.2008.11.030 |
| [7] |
KABIR E R, RAHMAN M S, RAHMAN I. A review on endocrine disruptors and their possible impacts on human health[J]. Environmental Toxicology and Pharmacology, 2015, 40(1): 241-258. DOI:10.1016/j.etap.2015.06.009 |
| [8] |
CHANG H S, CHOO K H, LEE B, et al. The methods of identification, analysis, and removal of endocrine disrupting compounds(EDCs) in water[J]. Journal of Hazardous Materials, 2009, 172(1): 1-12. DOI:10.1016/j.jhazmat.2009.06.135 |
| [9] |
GUO J, SHI W, CHEN Q, et al. Extended virtual screening strategies to link antiandrogenic activities and detected organic contaminants in soils[J]. Environmental Science & Technology, 2017, 51(21): 12528-12536. |
| [10] |
WU G, WANG X, ZHANG X, et al. Nontarget screening based on molecular networking strategy to identify transformation products of citalopram and sertraline in wastewater[J]. Water Research, 2023, 232: 119509. DOI:10.1016/j.watres.2022.119509 |
| [11] |
LI H, YI X, CHENG F, et al. Identifying organic toxicants in sediment using effect-directed analysis: A combination of bioaccessibility-based extraction and high-throughput midge toxicity testing[J]. Environmental Science & Technology, 2019, 53(2): 996-1003. |
| [12] |
RAHU I, KULL M, KRUVE A. Predicting the activity of unidentified chemicals in complementary bioassays from the HRMS data to pinpoint potential endocrine disruptors[J]. Journal of Chemical Information and Modeling, 2024, 64(8): 3093-3104. DOI:10.1021/acs.jcim.3c02050 |
| [13] |
JONKERS T J H, MEIJER J, VLAANDEREN J J, et al. High-per formance data processing workflow incorporating effect-directed analysis for feature prioritization in suspect and nontarget screening[J]. Environmental Science & Technology, 2022, 56(3): 1639-1651. |
| [14] |
HOLLENDER J, SCHYMANSKI E L, SINGER H P, et al. Nontarget screening with high resolution mass spectrometry in the environment: Ready to go?[J]. Environmental Science & Technology, 2017, 51(20): 11505-11512. |
| [15] |
SCHULZE B, JEON Y, KASERZON S, et al. An assessment of quality assurance/quality control efforts in high resolution mass spectrometry non-target workflows for analysis of environmental samples[J]. TrAC Trends in Analytical Chemistry, 2020, 133: 116063. DOI:10.1016/j.trac.2020.116063 |
| [16] |
LI Q, WANG L, JIA Y, et al. Nontargeted analysis reveals a broad range of bioactive pollutants in drinking water by estrogen receptor affinity-mass spectrometry[J]. Environmental Science & Technology, 2023, 57(50): 21327-21336. |
| [17] |
MOSCHET C, PIAZZOLI A, SINGER H, et al. Alleviating the reference standard dilemma using a systematic exact mass suspect screening approach with liquid chromatography-high resolution mass spectrometry[J]. Analytical Chemistry, 2013, 85(21): 10312-10320. DOI:10.1021/ac4021598 |
| [18] |
KESHET U, KIND T, LU X, et al. Acyl-CoA identification in mouse liver samples using the in silico CoA-blast tandem mass spectral library[J]. Analytical Chemistry, 2022, 94(6): 2732-2739. DOI:10.1021/acs.analchem.1c03272 |
| [19] |
SU H, REN K, LI R, et al. Suspect screening of liquid crystal monomers(LCMs) in sediment using an established database covering 1173 LCMs[J]. Environmental Science & Technology, 2022, 56(12): 8061-8070. |
| [20] |
FAN F, WU G, YANG Y, et al. A graph neural network model with a transparent decision-making process defines the applicability domain for environmental estrogen screening[J]. Environmental Science & Technology, 2023, 57(46): 18236-18245. |
| [21] |
GUO J, SHEN Y, ZHANG X, et al. Effect-directed analysis based on the reduced human transcriptome(RHT) to identify organic contaminants in source and tap waters along the yangtze river[J]. Environmental Science & Technology, 2022, 56(12): 7840-7852. |
| [22] |
WANG L, ZHAO L, LIU X, et al. SepPCNET: Deeping learning on a 3D surface electrostatic potential point cloud for enhanced toxicity classification and its application to suspected environmental estrogens[J]. Environmental Science & Technology, 2021, 55(14): 9958-9967. |
| [23] |
ZGHEIB E, KIM M J, JORNOD F, et al. Identification of non-validated endocrine disrupting chemical characterization methods by screening of the literature using artificial intelligence and by database exploration[J]. Environment International, 2021, 154: 106574. DOI:10.1016/j.envint.2021.106574 |
| [24] |
MUSCHKET M, DI PAOLO C, TINDALL A J, et al. Identification of unknown antiandrogenic compounds in surface waters by effect-directed analysis(EDA) using a parallel fractionation approach[J]. Environmental Science & Technology, 2018, 52(1): 288-297. |
| [25] |
BLACK G P, ANUMOL T, YOUNG T M. Analyzing a broader spectrum of endocrine active organic contaminants in sewage sludge with high resolution LC-QTOF-MS suspect screening and QSAR toxicity prediction[J]. Environmental Science: Processes & Impacts, 2019, 21(7): 1099-1114. |
| [26] |
LEUSCH F D L, NEALE P A, ARNAL C, et al. Analysis of endocrine activity in drinking water, surface water and treated wast-ewater from six countries[J]. Water Research, 2018, 139: 10-18. DOI:10.1016/j.watres.2018.03.056 |
| [27] |
LIU J, XIANG T, SONG X C, et al. High-efficiency effect-directed analysis leveraging five high level advancements: A critical review[J]. Environmental Science & Technology, 2024, 58(23): 9925-9944. |
| [28] |
DONG H, CUTHBERTSON A A, RICHARDSON S D. Effect-directed analysis(EDA): A promising tool for nontarget identification of unknown disinfection byproducts in drinking water[J]. Environmental Science & Technology, 2020, 54(3): 1290-1292. |
| [29] |
ALYGIZAKIS N, LESTREMAU F, GAGO-FERRERO P, et al. Towards a harmonized identification scoring system in LC-HRMS/MS based non-target screening(NTS) of emerging contaminants[J]. TrAC Trends in Analytical Chemistry, 2023, 159: 116944. DOI:10.1016/j.trac.2023.116944 |
| [30] |
FISHER C M, CROLEY T R, KNOLHOFF A M. Data processing strategies for non-targeted analysis of foods using liquid chromatography/high-resolution mass spectrometry[J]. TrAC Trends in Analytical Chemistry, 2021, 136: 116188. DOI:10.1016/j.trac.2021.116188 |
| [31] |
TSUGAWA H, CAJKA T, KIND T, et al. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis[J]. Nature Methods, 2015, 12(6): 523-526. DOI:10.1038/nmeth.3393 |
| [32] |
PLUSKAL T, CASTILLO S, VILLAR-BRIONES A, et al. MZmine 2:Modular framework for processing, visualizing, and analy-zing mass spectrometry-based molecular profile data[J]. BMC Bioinformatics, 2010, 11(1): 395. DOI:10.1186/1471-2105-11-395 |
| [33] |
TAUTENHAHN R, PATTI G J, RINEHART D, et al. XCMS online: A web-based platform to process untargeted metabolomic data[J]. Analytical Chemistry, 2012, 84(11): 5035-5039. DOI:10.1021/ac300698c |
| [34] |
HOHRENK L L, ITZEL F, BAETZ N, et al. Comparison of software tools for liquid chromatography-high-resolution mass spectrometry data processing in nontarget screening of environmental samples[J]. Analytical Chemistry, 2020, 92(2): 1898-1907. DOI:10.1021/acs.analchem.9b04095 |
| [35] |
AISPORNA A, BENTON H P, CHEN A, et al. Neutral loss mass spectral data enhances molecular similarity analysis in METLIN[J]. Journal of the American Society for Mass Spectrometry, 2022, 33(3): 530-534. DOI:10.1021/jasms.1c00343 |
| [36] |
TSUGAWA H, NAKABAYASHI R, MORI T, et al. A cheminformatics approach to characterize metabolomes in stable-isotope-labeled organisms[J]. Nature Methods, 2019, 16(4): 295-298. DOI:10.1038/s41592-019-0358-2 |
| [37] |
ZENG Z, LIU X, DAI W, et al. Ion fusion of high-resolution LC-MS-based metabolomics data to discover more reliable biomarkers[J]. Analytical Chemistry, 2014, 86(8): 3793-3800. DOI:10.1021/ac500878x |
| [38] |
KOUŘIL Š, DE SOUSA J, VÁCLAVÍK J, et al. CROP: Correlation-based reduction of feature multiplicities in untargeted metabolomic data[J]. Bioinformatics, 2020, 36(9): 2941-2942. DOI:10.1093/bioinformatics/btaa012 |
| [39] |
WATROUS J, ROACH P, ALEXANDROV T, et al. Mass spectral molecular networking of living microbial colonies[J]. Proceedings of the National Academy of Sciences, 2012, 109(26): 1743-1752. |
| [40] |
ZHOU Z, LUO M, ZHANG H, et al. Metabolite annotation from knowns to unknowns through knowledge-guided multi-layer metabolic networking[J]. Nature Communications, 2022, 13(1): 6656. DOI:10.1038/s41467-022-34537-6 |
| [41] |
WU G, YAO R, ZHANG Y, et al. Transformation mechanisms of antidepressants in biological wastewater treatment: Removal kinetic, transformation products and pathways[J]. Chemical Engineering Journal, 2024, 493: 152557. DOI:10.1016/j.cej.2024.152557 |
| [42] |
WU G, QIAN Y, FAN F, et al. Revealing specific transformation pattern of sulfonamides during wastewater biological treatment processes by molecular networking nontarget screening[J]. Water Research, 2023, 235: 119895. DOI:10.1016/j.watres.2023.119895 |
| [43] |
YANG J Y, SANCHEZ L M, RATH C M, et al. Molecular networking as a dereplication strategy[J]. Journal of Natural Products, 2013, 76(9): 1686-1699. DOI:10.1021/np400413s |
| [44] |
ZHANG X, LI Z, ZHAO C, et al. Leveraging unidentified metabolic features for key pathway discovery: Chemical classi-fication-driven network analysis in untargeted metabolomics[J]. Analytical Chemistry, 2024, 96(8): 3409-3418. DOI:10.1021/acs.analchem.3c04591 |
| [45] |
ZWART N, NIO S L, HOUTMAN C J, et al. High-throughput effect-directed analysis using downscaled in vitro reporter gene assays to identify endocrine disruptors in surface water[J]. Environmental Science & Technology, 2018, 52(7): 4367-4377. |
| [46] |
HOUTMAN C J, TEN BROEK R, VAN OORSCHOT Y, et al. High resolution effect-directed analysis of steroid hormone(ant)agonists in surface and wastewater quality monitoring[J]. Environmental Toxicology and Pharmacology, 2020, 80: 103460. DOI:10.1016/j.etap.2020.103460 |
| [47] |
HOUTMAN C J, BREWSTER K, TEN BROEK R, et al. Characterisation of(anti-)progestogenic and(anti-)androgenic activities in surface and wastewater using high resolution effectdirected analysis[J]. Environment International, 2021, 153: 106536. DOI:10.1016/j.envint.2021.106536 |
| [48] |
XING S, JIAO Y, SALEHZADEH M, et al. SteroidXtract: Deep learning-based pattern recognition enables comprehensive and rapid extraction of steroid-like metabolic features for automated biology-driven metabolomics[J]. Analytical Chemistry, 2021, 93(14): 5735-5743. DOI:10.1021/acs.analchem.0c04834 |
| [49] |
GUO H, XUE K, SUN H, et al. Contrastive learning-based embedder for the representation of tandem mass spectra[J]. Analytical Chemistry, 2023, 95(20): 7888-7896. DOI:10.1021/acs.analchem.3c00260 |
| [50] |
ZWART N, JONKER W, BROEK R T, et al. Identification of mutagenic and endocrine disrupting compounds in surface water and wastewater treatment plant effluents using high-resolution effect-directed analysis[J]. Water Research, 2020, 168: 115204. DOI:10.1016/j.watres.2019.115204 |
| [51] |
HASHMI M A K, KRAUSS M, ESCHER B I, et al. Effect‐directed analysis of progestogens and glucocorticoids at trace concentrations in river water[J]. Environmental Toxicology and Chemistry, 2020, 39(1): 189-199. DOI:10.1002/etc.4609 |
| [52] |
LOPEZ-HERGUEDAS N, GONZÁLEZ-GAYA B, CANO A, et al. Effect-directed analysis of a hospital effluent sample using a-YES for the identification of endocrine disrupting compounds[J]. Science of The Total Environment, 2022, 850: 157985. DOI:10.1016/j.scitotenv.2022.157985 |
| [53] |
HASHMI M A K, ESCHER B I, KRAUSS M, et al. Effect-directed analysis(EDA) of danube river water sample receiving untreated municipal wastewater from novi sad, serbia[J]. Science of The Total Environment, 2018, 624: 1072-1081. DOI:10.1016/j.scitotenv.2017.12.187 |
| [54] |
JANG I, LEE J U, LEE J M, et al. LC-MS/MS software for screening unknown erectile dysfunction drugs and analogues: Artificial neural network classification, peak-count scoring, simple similarity search, and hybrid similarity search algorithms[J]. Analytical Chemistry, 2019, 91(14): 9119-9128. DOI:10.1021/acs.analchem.9b01643 |
| [55] |
STEINBECK C, HAN Y, KUHN S, et al. The chemistry development kit(CDK): An open-source java library for chemo-and bioinformatics[J]. Journal of Chemical Information and Computer Sciences, 2003, 43(2): 493-500. DOI:10.1021/ci025584y |
 2024, Vol. 16
2024, Vol. 16 

